For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSETDLSPRRAPTSAQFVAMQASPEFKELRSRLRRFVFPMTAFFLVWYAIYVALGAFAHDFMAIQVIGNI NVGLIIGLGQFLSTFVITGLYVRFANRELDPRAAAIREELEGPAQ
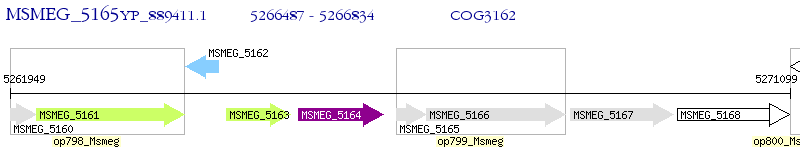
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5165 | - | - | 100% (115) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2140 | - | 1e-40 | 73.21% (112) | hypothetical protein Mflv_2140 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4563 | - | 2e-41 | 75.00% (112) | hypothetical protein Mvan_4563 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2140|M.gilvum_PYR-GCK MTYISTTFHPEPVHSSQEREIKVSETNLPIRPEAISGDRYLEVQASPEFQ
Mvan_4563|M.vanbaalenii_PYR-1 ----------------------MSETDLPIRPEAISGERYLEVQASPEFQ
MSMEG_5165|M.smegmatis_MC2_155 ----------------------MSETDLSPR-RAPTSAQFVAMQASPEFK
:***:*. * .* :. ::: :******:
Mflv_2140|M.gilvum_PYR-GCK ELRSRLRRFVFPMTAVFLVWYTTYVLLGAFATDFMATKVWGDVNVGLLIG
Mvan_4563|M.vanbaalenii_PYR-1 ELRTRLRRFVFPMTAVFLVWYAVYVLLGAFATGFMGTKVWGDINVGLIIG
MSMEG_5165|M.smegmatis_MC2_155 ELRSRLRRFVFPMTAFFLVWYAIYVALGAFAHDFMAIQVIGNINVGLIIG
***:***********.*****: ** ***** .**. :* *::****:**
Mflv_2140|M.gilvum_PYR-GCK LGQFLSTFLITGLYVRFANRELDPRAEAIRTQLESEIR
Mvan_4563|M.vanbaalenii_PYR-1 LGQFLSTFLITGLYVRFANRELDPRAAAIRTQLESEIR
MSMEG_5165|M.smegmatis_MC2_155 LGQFLSTFVITGLYVRFANRELDPRAAAIREELEGPAQ
********:***************** *** :**. :