For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VELLTGFGLATAAGLNAYIPLLALGLLSRFTDLVALPAGWAWLENGWVMAIVAVLLVVEIVADKIPALDT VNDTIQTFVRPTAGGIVFGSGTAAETAAVTDPGAFAQSGQWIPVAIGVVTALVVSLTKTAVRPAANVASA GVAAPVLSTVEDVTSVGLVIVAILLPVLVLVALVLMAWGAFRLWRWRRRRAKSTHPSPPQPLPPR
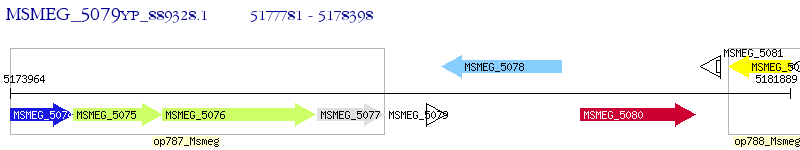
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5079 | - | - | 100% (205) | hypothetical protein MSMEG_5079 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2195 | - | 3e-86 | 81.87% (193) | hypothetical protein Mflv_2195 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3873c | - | 3e-29 | 38.46% (195) | hypothetical protein MAB_3873c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4502 | - | 1e-86 | 83.68% (190) | hypothetical protein Mvan_4502 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2195|M.gilvum_PYR-GCK -------MELLTGFGLATAAGLNAYIPLLALGLLGRFTELVTLPAGWAWL
Mvan_4502|M.vanbaalenii_PYR-1 -------MELLTGFGLATAAGLNAYIPLLTLGLLARFTDLVTLPAGWAWL
MSMEG_5079|M.smegmatis_MC2_155 -------MELLTGFGLATAAGLNAYIPLLALGLLSRFTDLVALPAGWAWL
MAB_3873c|M.abscessus_ATCC_199 MEITSAISAILGAFGLSGAAGLNAWLPLLAVGAADRLG-WIELGPSYGWL
:* .***: ******::***::* *: : * ..:.**
Mflv_2195|M.gilvum_PYR-GCK ENGWVMTIVAALLVIEVVADKVPALDSVNDVVQTFVRPTAGGIVFGSGTA
Mvan_4502|M.vanbaalenii_PYR-1 ENGWVMAIVAVLLVVEVVADKVPALDSVNDVIQTFVRPTAGGIVFGSGTA
MSMEG_5079|M.smegmatis_MC2_155 ENGWVMAIVAVLLVVEIVADKIPALDTVNDTIQTFVRPTAGGIVFGSGTA
MAB_3873c|M.abscessus_ATCC_199 SSTPALVVLAVVFVLDLIGDKVPALDSVLHAIGAFIAPASGAVLFTAETS
.. .:.::*.::*::::.**:****:* ..: :*: *::*.::* : *:
Mflv_2195|M.gilvum_PYR-GCK AQTATVTDPGAFAQSGQWIPVAIGVAIALVVSLTKSTVRPAANVATAGVA
Mvan_4502|M.vanbaalenii_PYR-1 AQTAAVTDPGAFAQTGQWVPVVIGVVVALVVSLTKSTVRPAANVATAGVA
MSMEG_5079|M.smegmatis_MC2_155 AETAAVTDPGAFAQSGQWIPVAIGVVTALVVSLTKTAVRPAANVASAGVA
MAB_3873c|M.abscessus_ATCC_199 LSSNLPPA----------VAAILGAITAGGVHASRTVARPFVTGTTAGVG
.: . :.. :*. * * :::..** .. ::***.
Mflv_2195|M.gilvum_PYR-GCK APVLSTVEDVASVALVFVAILVPVL----VLVAVLALAWAAVRLLRRRRA
Mvan_4502|M.vanbaalenii_PYR-1 APVLSTVEDIVSVGLAFIAILLPVL----VLVVAGALAWAAVRLIRRRRA
MSMEG_5079|M.smegmatis_MC2_155 APVLSTVEDVTSVGLVIVAILLPVL----VLVALVLMAWGAFRLWRWRRR
MAB_3873c|M.abscessus_ATCC_199 NPVVSTAEDGTSLVLTILALAVPILAFLVVLVLLAGLGWLTYRAIRWARG
**:**.** .*: *.::*: :*:* *** :.* : * * *
Mflv_2195|M.gilvum_PYR-GCK AAQTPRN---------
Mvan_4502|M.vanbaalenii_PYR-1 RQAT------------
MSMEG_5079|M.smegmatis_MC2_155 RAKSTHPSPPQPLPPR
MAB_3873c|M.abscessus_ATCC_199 RRESDATDG-------
: