For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSQEPDPSPVTNSDSAPTEPTFADLQIHPAVLQAVTDVGYESPSAIQAATIPPMLAGSDVVGLAQTGTGK TAAFAIPILSKIDTSRRDTQALVLAPTRELALQVAEAFGRYGSHLPQVNVLPIYGGSSYTVQLSGLRRGA QVVVGTPGRVIDHLERGTLDLSKLDYLVLDEADEMLTMGFAEEVERILADTPEYKQVALFSATMPAAIRK ITTKYLHDPVEVTVKAKTATAENISQRFIQVAGPRKMDALTRILEVEEGDAMIVFVRTKQATEEVADRLK ARGFAAAAINGDINQAQRERTISALKDGTIDILIATDVAARGLDVERISHVINYDIPHDTESYVHRIGRT GRAGRSGHAVLFVTPRERHLLKSIEKATRSKLIEAELPSVEDVNAQRVAKFRDSISDSLNAPGIDLFRRL IEDYERESNVPLADIAAALAVQTRNGEEFLMTEPPPDKRRDRDRDRDDRGDRGPRKERPPREGLATYRID VGKRHKVGPGHIVGAIANEGGLHRNEFGHITIRLDYSLVELPEKLPKKTLKALENTRISGVLINLQPDRG PRREGAGKPSGKYKPHRKVK*
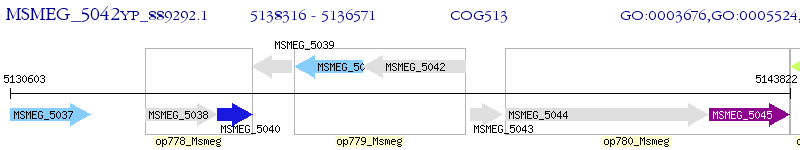
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5042 | - | - | 100% (581) | ATP-dependent rna helicase, dead/deah box family protein |
| M. smegmatis MC2 155 | MSMEG_1930 | - | 5e-81 | 43.36% (369) | DEAD/DEAH box helicase |
| M. smegmatis MC2 155 | MSMEG_1540 | - | 2e-60 | 34.16% (363) | ATP-dependent RNA helicase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1285 | deaD | 0.0 | 74.91% (562) | cold-shock DEAD-box protein A |
| M. gilvum PYR-GCK | Mflv_2225 | - | 0.0 | 79.07% (559) | DEAD/DEAH box helicase domain-containing protein |
| M. tuberculosis H37Rv | Rv1253 | deaD | 0.0 | 74.73% (562) | cold-shock DEAD-box protein A |
| M. leprae Br4923 | MLBr_00811 | rhlE | 4e-82 | 42.30% (383) | putative ATP-dependent RNA helicase |
| M. abscessus ATCC 19977 | MAB_1403 | - | 0.0 | 78.78% (575) | cold-shock DEAD box protein A homolog (ATP-dependent RNA |
| M. marinum M | MMAR_4189 | deaD | 0.0 | 76.39% (576) | cold-shock DEAD-box protein, DeaD |
| M. avium 104 | MAV_1401 | - | 0.0 | 78.23% (565) | ATP-dependent rna helicase, dead/deah box family protein |
| M. thermoresistible (build 8) | TH_2231 | - | 0.0 | 82.24% (563) | ATP-dependent rna helicase, dead/deah box family protein |
| M. ulcerans Agy99 | MUL_2533 | rhlE | 4e-79 | 41.88% (382) | ATP-dependent RNA helicase RhlE |
| M. vanbaalenii PYR-1 | Mvan_4470 | - | 0.0 | 80.11% (563) | DEAD/DEAH box helicase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5042|M.smegmatis_MC2_155 -----------MTSQEPDPSPVTNSDSAPTEPTFADLQIHPAVLQAVTDV
TH_2231|M.thermoresistible__bu ---------------------VSG-----TDPTFADLQIHPAVLRAVGDV
Mflv_2225|M.gilvum_PYR-GCK -------------------MTSSNAEPDPADLTFADLQIHPAVLKAVADV
Mvan_4470|M.vanbaalenii_PYR-1 -------------------MTSSNAETESADLTFADLQIHPAVLQAVADV
Mb1285|M.bovis_AF2122/97 -------------------MAFPEYSPAASAATFADLQIHPRVLRAIGDV
Rv1253|M.tuberculosis_H37Rv -------------------MAFPEYSPAASAATFADLQIHPRVLRAIGDV
MMAR_4189|M.marinum_M -------------------MANPENPPAVPPATFADLQIHPSVMRAIADV
MAV_1401|M.avium_104 -------------------MTLPDSSTEAASPTFADLQIHPSVLRAIADV
MAB_1403|M.abscessus_ATCC_1997 ---------MTSTDIPTPDTGDSSAPPADDTVTFADLHIAPEVLRAVSDV
MLBr_00811|M.leprae_Br4923 MNRDSKTPTQNRFSHRKAFHPHMTAPTSTAEPAFAKLGVRDEIVRALAER
MUL_2533|M.ulcerans_Agy99 -------------------MTALKSTTETTPKTFAELGVRDEIVRALGER
:**.* : :::*: :
MSMEG_5042|M.smegmatis_MC2_155 GYESPSAIQAATIPPMLAGSDVVGLAQTGTGKTAAFAIPILSKIDTSR--
TH_2231|M.thermoresistible__bu GYETPSAIQAATIPAMMAGSDVVGLAQTGTGKTAAFAIPILSKIDTAS--
Mflv_2225|M.gilvum_PYR-GCK GYESPSPIQAATIPAILEGSDVVGLAQTGTGKTAAFAIPILSKIDTDS--
Mvan_4470|M.vanbaalenii_PYR-1 GYETPSPIQAATIPAILDGSDVVGLAQTGTGKTAAFAIPILSKIDTDS--
Mb1285|M.bovis_AF2122/97 GYESPTAIQAATIPALMAGSDVVGLAQTGTGKTAAFAIPMLSKIDITS--
Rv1253|M.tuberculosis_H37Rv GYESPTAIQAATIPALMAGSDVVGLAQTGTGKTAAFAIPMLSKIDITS--
MMAR_4189|M.marinum_M GYESPTGIQAATIPALMAGSDVVGLAQTGTGKTAAFAIPILSKIDTTS--
MAV_1401|M.avium_104 GYETPTGIQAATIPALMAGSDVVGLAQTGTGKTAAFAIPILSKIDVAS--
MAB_1403|M.abscessus_ATCC_1997 GYETPSAIQAATIPPLLAGSDVVGLAQTGTGKTAAFAIPILSKIDLTS--
MLBr_00811|M.leprae_Br4923 GIQQPFAIQELTLPLALAGDDVIGQARTGMGKTFAFGVPLLQRISVTTTA
MUL_2533|M.ulcerans_Agy99 DIEGPFAIQELTLPLALAGDDLIGQARTGMGKTFAFGVPLLQRVSSGTGV
. : * ** *:* : *.*::* *:** *** **.:*:*.::.
MSMEG_5042|M.smegmatis_MC2_155 ----RDTQALVLAPTRELALQVAEAFGRYG---------SHLPQVNVLPI
TH_2231|M.thermoresistible__bu ----RATQALVLAPTRELALQVAEAFGRYG---------AHLPDVQVLPI
Mflv_2225|M.gilvum_PYR-GCK ----RNTQALVLAPTRELALQVAEAFGRYG---------AQLR-VNVLPI
Mvan_4470|M.vanbaalenii_PYR-1 ----RNTQALVLAPTRELALQVAEAFGRYG---------AHLR-VNVLPI
Mb1285|M.bovis_AF2122/97 ----KVPQALVLVPTRELALQVAEAFGRYG---------AYLSQLNVLPI
Rv1253|M.tuberculosis_H37Rv ----KVPQALVLVPTRELALQVAEAFGRYG---------AYLSQLNVLPI
MMAR_4189|M.marinum_M ----TATQALVLAPTRELALQVAEAFSRYG---------AHLSRLNVLPV
MAV_1401|M.avium_104 ----TATQALVLAPTRELALQVAEAFSRYG---------AHLPKINVLPI
MAB_1403|M.abscessus_ATCC_1997 ----KDTQALVLAPTRELALQVAEAFGRYG---------AHMPKLNVLPI
MLBr_00811|M.leprae_Br4923 RPLSGAPRGLVVVPTRELCLQVADDLATVAKYLTAD---DDNRRFSVVSI
MUL_2533|M.ulcerans_Agy99 RPLTGAPRALVVVPTRELCLQVTEDLAVAAKYLTATTETGDQRPLSVVSI
.:.**:.*****.***:: :. . ..*:.:
MSMEG_5042|M.smegmatis_MC2_155 YGGSSYTVQLSGLRRGAQVVVGTPGRVIDHLERGTLDLSKLDYLVLDEAD
TH_2231|M.thermoresistible__bu YGGSSYTVQLSGLRRGAQIVVGTPGRVIDHLDRGTLDLSRLDYLVLDEAD
Mflv_2225|M.gilvum_PYR-GCK YGGSSYVPQLAGLKRGAQVVVGTPGRVIDHLEKGSLDLSHLDYLVLDEAD
Mvan_4470|M.vanbaalenii_PYR-1 YGGASYVPQLSGLKRGAQVVVGTPGRVIDHLEKGSLDLSHLDYLVLDEAD
Mb1285|M.bovis_AF2122/97 YGGSSYAVQLAGLRRGAQVVVGTPGRVIDHLERATLDLSRVDFLVLDEAD
Rv1253|M.tuberculosis_H37Rv YGGSSYAVQLAGLRRGAQVVVGTPGRMIDHLERATLDLSRVDFLVLDEAD
MMAR_4189|M.marinum_M YGGSSYAIQLAGLRRGAQVVVGTPGRVIDHLERRTLDLSRVDYLVLDEAD
MAV_1401|M.avium_104 YGGSSYAVQLAGLKRGAHVVVGTPGRVIDHLERGTLDLSHVDYLVLDEAD
MAB_1403|M.abscessus_ATCC_1997 YGGQSYVVQLAGLKRGAQVVVGTPGRVIDHLERGTLDLSHLDYLVLDEAD
MLBr_00811|M.leprae_Br4923 YGGRAYEPQIEALQTGADVVVSTPGRLLDLCQQGHLQLSGLSVLVLDEAD
MUL_2533|M.ulcerans_Agy99 YGGRAYEPQIEALRAGADVVVGTPGRLLDLSQQGHLQLGGLYVLVLDEAD
*** :* *: .*: **.:**.****::* :: *:*. : *******
MSMEG_5042|M.smegmatis_MC2_155 EMLTMGFAEEVERILADTPEYKQVALFSATMPAAIRKITTKYLHDPVEVT
TH_2231|M.thermoresistible__bu EMLQMGFAEDVERILADTPEYKQVALFSATMPAAIRKLTTKYLHDPVEVT
Mflv_2225|M.gilvum_PYR-GCK EMLQMGFAEDVERILADTPEYKQVALFSATMPPAIKKITAKYLHDPVEVT
Mvan_4470|M.vanbaalenii_PYR-1 EMLQMGFAEDVERILADTPEYKQVALFSATMPPAIKKITAKYLHDPVEVT
Mb1285|M.bovis_AF2122/97 EMLTMGFADDVERILSETPEYKQVALFSATMPPAIRKLSAKYLHDPFEVT
Rv1253|M.tuberculosis_H37Rv EMLTMGFADDVERILSETPEYKQVALFSATMPPAIRKLSAKYLHDPFEVT
MMAR_4189|M.marinum_M EMLTMGFAEDVERILSETPEYKQVALFSATMPPAIRKITTRYLHDPLEVK
MAV_1401|M.avium_104 EMLTMGFAEEVDRILSETPEYKQVALFSATMPPAIRKLTAKYLHDPLEVS
MAB_1403|M.abscessus_ATCC_1997 EMLTMGFAEDVERILADTPEYKQVALFSATMPTTIRRLTKKYLHDPVEIK
MLBr_00811|M.leprae_Br4923 EMLDLGFFPDIERLLGQIPTDRQSMLFSATMPDPIITLARTFMDQPIHIR
MUL_2533|M.ulcerans_Agy99 EMLDLGFLPDIERILRQIPADRQSMLFSATMPDPIITLARTFMIQPTHIR
*** :** :::*:* : * :* ******* .* :: :: :* .:
MSMEG_5042|M.smegmatis_MC2_155 VKAKT--ATAENISQRFIQVAGPRKMDALTRILEVEEGDAMIVFVRTKQA
TH_2231|M.thermoresistible__bu VKART--ATAENITQRFIQVAGPRKMDALTRVLETEPFEAMIVFVRTKQA
Mflv_2225|M.gilvum_PYR-GCK VKSKT--QTAENITQRYFLVSYPRKMDALTRLLETEQGDAMIVFVRTKQA
Mvan_4470|M.vanbaalenii_PYR-1 VKSKT--QTAENITQRYFLVSYPRKMDALTRLLEVEQGDAMIVFVRTKQA
Mb1285|M.bovis_AF2122/97 CKAKT--AVAENISQSYIQVA--RKMDALTRVLEVEPFEAMIVFVRTKQA
Rv1253|M.tuberculosis_H37Rv CKAKT--AVAENISQSYIQVA--RKMDALTRVLEVEPFEAMIVFVRTKQA
MMAR_4189|M.marinum_M SEAKT--ATAENISQRYIQVAGPRKMDALTRVLEVEPFEAMIVFVRTKQA
MAV_1401|M.avium_104 TKAKT--TTAENISQRYIQVAGPRKMDALTRVLEVEPFEAMIVFVRTKQA
MAB_1403|M.abscessus_ATCC_1997 IDAKT--STAENITQRFIQVAGPRKMDALTRILEVETFEAMIVFVRTKQA
MLBr_00811|M.leprae_Br4923 AEAPHSLAVHDTTQQFVYRAHALDKVELISRILQARGRGATMIFTRTKRT
MUL_2533|M.ulcerans_Agy99 AEAPHSLAVHDTTEQFVYRAHALDKVELVTRVLQARERGATMIFTRTKRT
.: . :. * . *:: ::*:*:.. * ::*.***::
MSMEG_5042|M.smegmatis_MC2_155 TEEVADRLKARGFAAAAINGDINQAQRERTISALKDGTIDILIATDVAAR
TH_2231|M.thermoresistible__bu TEELAERLRSRGFAAAAINGDIAQAQRERTIAALKDGSLDILIATDVAAR
Mflv_2225|M.gilvum_PYR-GCK TEEVAEKLRARGFAASAINGDIPQAVRERTIAQLKDGTIDILVATDVAAR
Mvan_4470|M.vanbaalenii_PYR-1 TEEVAEKLRSRGFAAAAINGDIPQAVRERTISQLKDGTIDILVATDVAAR
Mb1285|M.bovis_AF2122/97 TEEIAEKLRARGFSAAAISGDVPQAQRERTITALRDGDIDILVATDVAAR
Rv1253|M.tuberculosis_H37Rv TEEIAEKLRARGFSAAAISGDVPQAQRERTITALRDGDIDILVATDVAAR
MMAR_4189|M.marinum_M TEEVAEKLRARGFSAAAINGDIPQGQRERTITALRDGGIDILVATDVAAR
MAV_1401|M.avium_104 TEEVAERLRARGFSAAAINGDIPQGQRERTVAALKDGGIDILVATDVAAR
MAB_1403|M.abscessus_ATCC_1997 TEEVADKLKARGFAAAAINGDINQSQRERTINALKAGTIDILVATDVAAR
MLBr_00811|M.leprae_Br4923 AQKVTNELNERGFAVGAVHGDLGQIARENALEAFRTGNVDVLVATDVAAR
MUL_2533|M.ulcerans_Agy99 AQKVADELNERGFAVGAVHGDLGQVAREKALKSFRRGSVNVLVATDVAAR
::::::.*. ***:..*: **: * **.:: :: * :::*:*******
MSMEG_5042|M.smegmatis_MC2_155 GLDVERISHVINYDIPHDTESYVHRIGRTGRAGRSGHAVLFVTPRERHLL
TH_2231|M.thermoresistible__bu GLDVDRISHVVNYDIPHDTESYVHRIGRTGRAGRSGNALLFVTPRERHLL
Mflv_2225|M.gilvum_PYR-GCK GLDVERISHVVNFDIPHDPESYVHRIGRTGRAGRSGTALLFVTPRERHLL
Mvan_4470|M.vanbaalenii_PYR-1 GLDVERISHVVNFDIPHDPESYVHRIGRTGRAGRSGTALLFVTPRERHLL
Mb1285|M.bovis_AF2122/97 GLDVERISHVLNYDIPHDTESYVHRIGRTGRAGRSGAALIFVSPRELHLL
Rv1253|M.tuberculosis_H37Rv GLDVERISHVLNYDIPHDTESYVHRIGRTGRAGRSGAALIFVSPRELHLL
MMAR_4189|M.marinum_M GLDVERISHVLNYDIPHDTESYVHRIGRTGRAGRSGAALLFVSPRERHLL
MAV_1401|M.avium_104 GLDVERISHVLNYDIPHDTESYVHRIGRTGRAGRSGTALLFVSPRERHLL
MAB_1403|M.abscessus_ATCC_1997 GLDVERISHVLNYDIPHDTESYVHRIGRTGRAGRSGTAVLFVSPRERHLL
MLBr_00811|M.leprae_Br4923 GIDIDDVTHVINYQCPEDEKMYVHRIGRTGRAGRTGVAVTLVDWDELHRW
MUL_2533|M.ulcerans_Agy99 GIDIDDVTHVINYQCPEDEKMYVHRIGRTGRAGRTGVAVTLVDWDELPRW
*:*:: ::**:*:: *.* : *************:* *: :* *
MSMEG_5042|M.smegmatis_MC2_155 KSIEKATRSKLIEAELPSVEDVNAQRVAKFRDSISDSLNAPGIDLFRRLI
TH_2231|M.thermoresistible__bu KAIEKHTRSKVLEAEMPTVDDVNAKRVAKFRDAISEALSSPQIDLFRGLI
Mflv_2225|M.gilvum_PYR-GCK GAIERVTRQKLVESELPSVDDVNEKRVAKFRDSITDALDKPGIDLFRTLV
Mvan_4470|M.vanbaalenii_PYR-1 GAIERVTRQKLVESELPSVEDVNEKRVAKFRDSITDALEKPGIDLFRTLI
Mb1285|M.bovis_AF2122/97 KAIEKATRQTLTEAQLPTVEDVNTQRVAKFADSITNALGGPGIELFRRLV
Rv1253|M.tuberculosis_H37Rv KAIEKATRQTLTEAQLPTVEDVNTQRVAKFADSITNALGGPGIELFRRLV
MMAR_4189|M.marinum_M KLIEKATRQTLTEAELPTVEDVNAQRVAKFADSITHALSNTGIELFRRLV
MAV_1401|M.avium_104 KAIEKATRQPLTEAELPTVEDVNAQRVAKFADSITAALGAPGIDLFRKLV
MAB_1403|M.abscessus_ATCC_1997 KAIEKTTGAKLAEEPLPSVEDVNAQRVTKFRDAITAALAAPEVELFRRLV
MLBr_00811|M.leprae_Br4923 ETIDKALGLGTPDPAETYSNSPHLYTELAIPAEASGTVGTPRKAQVKRRK
MUL_2533|M.ulcerans_Agy99 TMIDKALGLGSPDPAETYSSSPHLYTELSIPTDAAGTVGPPSKSGSKPRT
*:: : . .. : : : :: . :
MSMEG_5042|M.smegmatis_MC2_155 EDYERESNVPLADIAAALAVQTRNGEEFLMTEPPPDKRRDRDRDRDDRGD
TH_2231|M.thermoresistible__bu EDYERDHDVPLADIAAALAVQTRDGDEFLMVEPPPEQRRERSERRDGR--
Mflv_2225|M.gilvum_PYR-GCK EGYERDHDVPMADIAAALALQSRNGEEFLMTEPPPEKRRER-PERG----
Mvan_4470|M.vanbaalenii_PYR-1 EGYERDHDVPMADIAAALALQSRNGEEFLMTEPPPEKRRER-RERDGR-D
Mb1285|M.bovis_AF2122/97 EEYEREHDVPMADIAAALAVQCRGGEAFLMAPDPPLSRRNRDQ-RRDR--
Rv1253|M.tuberculosis_H37Rv EEYEREHDVPMADIAAALAVQCRGGEAFLMAPDPPLSRRNRDQ-RRDR--
MMAR_4189|M.marinum_M EDYEREHNVPMADIAAALALQSRDGEEFLLAPDPPPDQRERHP-RNER--
MAV_1401|M.avium_104 QDYEREHDVPMADIAAALAVQSRDGEEFLMAPEPPRERRERHTERRER--
MAB_1403|M.abscessus_ATCC_1997 EDYERDNNVPVADIAAALAVLSRDGEQFLLQPDPPREPRHERGERPDR--
MLBr_00811|M.leprae_Br4923 TDRDGDKSGTSAQAAKAARTSNSVQRHRIRGGQP----------------
MUL_2533|M.ulcerans_Agy99 TEKQDDQQGEKSGRAR----SDAGRRRRTRGGKP----------------
: : . : * *
MSMEG_5042|M.smegmatis_MC2_155 RGPRKERPPREG----LATYRIDVGKRHKVGPGHIVGAIANEGGLHRNEF
TH_2231|M.thermoresistible__bu --ERRDRPPRRGDQ-KMATYRIAVGKRHKVGPGHIVGAIANEGGLHRSDF
Mflv_2225|M.gilvum_PYR-GCK --DASPRKPRERRS-DLATYRIAVGKRHKVAPGAIVGAIANEGGLHRSDF
Mvan_4470|M.vanbaalenii_PYR-1 GDDRGPRKPRERRS-DLATYRIAVGKRHKVAPGAIVGAIANEGGLHRSDF
Mb1285|M.bovis_AF2122/97 ----PQRPKRRPD---LTTYRVAVGKRHKIGPGAIVGAIANEGGLHRSDF
Rv1253|M.tuberculosis_H37Rv ----PQRPKRRPD---LTTYRVAVGKRHKIGPGAIVGAIANEGGLHRSDF
MMAR_4189|M.marinum_M ----KERPERRRSGGAFSTYRISVGKRHKVGPGAIVGAIANEGGLHRSDF
MAV_1401|M.avium_104 ----TEKPRSTRP---LATYRIAVGKRHKIGPGAIVGAIANEGGLHRSDF
MAB_1403|M.abscessus_ATCC_1997 ----GPRGERPRTP-GLATYRIAVGKRHKVMPGAIVGAIANEGGLHRSDF
MLBr_00811|M.leprae_Br4923 ----------------VTGHPTNNPSADSLATNGKVVAETLPNSSSGSPS
MUL_2533|M.ulcerans_Agy99 ----------------VTGHPATTPVAS-----GETATETGSGFGASSPS
.: : . : : . .
MSMEG_5042|M.smegmatis_MC2_155 GHITIRLDYSLVELPEKLPKKTLKALENTRISGVLINLQPDRGPRRE---
TH_2231|M.thermoresistible__bu GHITIRADYSLVELPAKLPRKALKALEHTRISGVPIDLRLDRGP-AH---
Mflv_2225|M.gilvum_PYR-GCK GHITIKMDHSLVELPAKLPGKTLKALQNTKIQGQLIQLQPDRGA------
Mvan_4470|M.vanbaalenii_PYR-1 GHITIKVDYSLVELPAKLSHKTLKALENTRIQGQLIHLEPDRGP------
Mb1285|M.bovis_AF2122/97 GQIRIGPDFSLVELPAKLPRATLKKLAQTRISGVLIDLRPYRPPD-----
Rv1253|M.tuberculosis_H37Rv GQIRIGPDFSLVELPAKLPRATLKKLAQTRISGVLIDLRPYRPPD-----
MMAR_4189|M.marinum_M GHITIGPDFSLVELPVKLPRETLKKLEQTRISGVLINLRPDRPADKA---
MAV_1401|M.avium_104 GHIAIGPDFSLVELPAKLPKSTLKRLEQTRISGVLINLQPDRAAA-----
MAB_1403|M.abscessus_ATCC_1997 GHITIRPDHSLVELPAKLSPKTLKALENTRISGVLIDLRPAGGPDGARPD
MLBr_00811|M.leprae_Br4923 TN---VRRRRHRRKPTEVAENGLDKNLAVQAN------------------
MUL_2533|M.ulcerans_Agy99 GNAGATRRRRRRRKPAGVQDN-----AAASAN------------------
: . * : . .
MSMEG_5042|M.smegmatis_MC2_155 -------------GAGKPSGKYKPHRKVK--
TH_2231|M.thermoresistible__bu -------------GPNRGPGRGGGKRKDR--
Mflv_2225|M.gilvum_PYR-GCK -----------------KPHRGKPPTQ----
Mvan_4470|M.vanbaalenii_PYR-1 -----------------KPHRGRPQSK----
Mb1285|M.bovis_AF2122/97 --------------AARRHNGGKPRRKHVG-
Rv1253|M.tuberculosis_H37Rv --------------AARRHNGGKPRRKHVG-
MMAR_4189|M.marinum_M -------------YKARRNDGGKPRRKRLG-
MAV_1401|M.avium_104 --------------KARGRDGGKPRRKYGG-
MAB_1403|M.abscessus_ATCC_1997 RKYTERRRADHKHSDHKRSEAKRPKRAPKKP
MLBr_00811|M.leprae_Br4923 -------------------------------
MUL_2533|M.ulcerans_Agy99 -------------------------------