For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNGQWLPDPEGRYEYRWWDGQSWTDQVSHQGQLHRAPLGPPPGPGPAQAQPQPQGDGFAGISGELVDGRF SEKEATAIANQNRKMLRVRLGEPFMARQGSMVAYQGNVDFAFEGGGASKFIKKALTGEGLPLMRCQGQGD VFLADRSFDVHLLQLTNTALSVSGKNVLAFSSSLDWNIERVRGGSMVAGGLFNTMLRGTGWVALTTDGPP VVLDASEAPTFADTNAVVAWSANLQTQLKMSFKAGALIGRGSGEAMQVAFHGNGFVIVQPSEGVTYPVQ
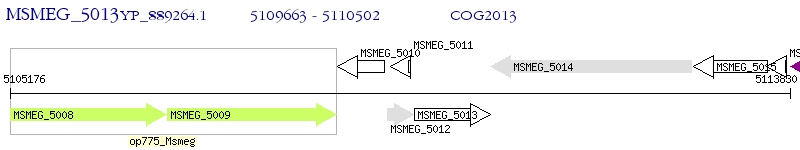
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5013 | - | - | 100% (279) | hypothetical protein MSMEG_5013 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2247 | - | 1e-133 | 83.70% (276) | hypothetical protein Mflv_2247 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4630 | - | 1e-124 | 78.10% (274) | hypothetical protein MAB_4630 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4445 | - | 1e-134 | 80.97% (289) | hypothetical protein Mvan_4445 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5013|M.smegmatis_MC2_155 ----------------MNG----QWLPDPEGRYEYRWWDGQSWTDQVSHQ
Mflv_2247|M.gilvum_PYR-GCK MTTHSVRSTDSSKEAAMTGPGQGSWQPDPEGRFDYRWHDGARWTDQVSHQ
Mvan_4445|M.vanbaalenii_PYR-1 ----------------MTGPGQGSWQPDPEGRFDYRWHDGQQWTDQVSHQ
MAB_4630|M.abscessus_ATCC_1997 ----------------MTTPNSGSWQPDPDGRYEFRWHDGQRWTDQVAHQ
*. .* ***:**:::** ** *****:**
MSMEG_5013|M.smegmatis_MC2_155 GQLHRAPLG-PPPGPGPAQA-------------QPQPQ---GDGFAGISG
Mflv_2247|M.gilvum_PYR-GCK GQIQRSPLGGAPPAQHPGQQ-------------QAQAVTGGGDGFTGITG
Mvan_4445|M.vanbaalenii_PYR-1 GQVHRAPFGGAPAPQAQQQAPQHQAPQHQAAQYQAQPVPGGGDGFAGISG
MAB_4630|M.abscessus_ATCC_1997 GSVSTAPLGGAAAQPAAPDQ-------------------PAGDQFSGISG
*.: :*:* ... : ** *:**:*
MSMEG_5013|M.smegmatis_MC2_155 ELVDGRFSEKEATAIANQNRKMLRVRLGEPFMARQGSMVAYQGNVDFAFE
Mflv_2247|M.gilvum_PYR-GCK DLVDGRFSEKEAKAIANQNTKLLRVRLGEPFMARQGSMVAYQGNVEFAFE
Mvan_4445|M.vanbaalenii_PYR-1 ELVDGRFSEKEAKPIANQNSKLLRVRLGEPFMARQGSMVAYQGNVDFAFE
MAB_4630|M.abscessus_ATCC_1997 DLVDGRFSENEAKPISNQNAKLLRVRLGEPFLARQGSMVAYQGSVDFAFE
:********:**..*:*** *:*********:***********.*:****
MSMEG_5013|M.smegmatis_MC2_155 GGGASKFIKKALTGEGLPLMRCQGQGDVFLADRSFDVHLLQLTNTALSVS
Mflv_2247|M.gilvum_PYR-GCK GGGASKFIKKALTGEGLPLMRCQGQGDVFLADRSYDVHLLNLNNSGLSIS
Mvan_4445|M.vanbaalenii_PYR-1 GGGASKFIKKALTGEGLPLMRCAGQGDVFLAERAYDVHLLNLTNSGLSIS
MAB_4630|M.abscessus_ATCC_1997 GGGAAKFLKKALTGEGLPLMRCSGQGDVFLAERAFDVHLLNLNNAGLSIS
****:**:************** ********:*::*****:*.*:.**:*
MSMEG_5013|M.smegmatis_MC2_155 GKNVLAFSSSLDWNIERVRGGSMVAGGLFNTMLRGTGWVALTTDGPPVVL
Mflv_2247|M.gilvum_PYR-GCK GKNVLAFSSSLDWNIERVKGGSMVAGGLFNTTLRGTGWVALTTDGPPVVL
Mvan_4445|M.vanbaalenii_PYR-1 GKNVLAFSSSLDWNIERVKGGSMVAGGLFNTTLRGTGWVALTTDGPPVVL
MAB_4630|M.abscessus_ATCC_1997 GKNVLAFSAGLNWNIERVRGASVVTGGLFNTTLRGTGWVALTTDGPPVVL
********:.*:******:*.*:*:****** ******************
MSMEG_5013|M.smegmatis_MC2_155 DASEAPTFADTNAVVAWSANLQTQLKMSFKAGALIGRGSGEAMQVAFHGN
Mflv_2247|M.gilvum_PYR-GCK NAAEAPTFADTNAVVAWSANLQTQLKTSFKAGALIGRGSGEAVQVSFYGN
Mvan_4445|M.vanbaalenii_PYR-1 NAAEAPTFADTNAVVAWSANLQTQLKTSFKAGALIGRGSGEAMQVSFYGE
MAB_4630|M.abscessus_ATCC_1997 NAAEAPTFADTNAVVAWSSNLQTQLKTSFKAGALIGRGSGEALQVSFQGR
:*:***************:******* ***************:**:* *.
MSMEG_5013|M.smegmatis_MC2_155 GFVIVQPSEGVTYPVQ-
Mflv_2247|M.gilvum_PYR-GCK GFVIVQPSEGIPVTTPQ
Mvan_4445|M.vanbaalenii_PYR-1 GFVIVQPSEGIQVTTAG
MAB_4630|M.abscessus_ATCC_1997 GFVIVQPSEGIPVVTAG
**********: .