For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DLDPNAIITAGALIGGGLIMGGGAIGAGIGDGIAGNALISGIARQPEAQGRLFTPFFITVGLVEAAYFIN LAFMALFVFATPGLQ*
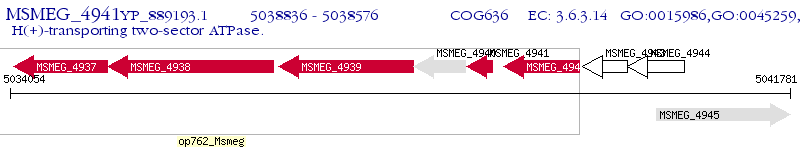
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4941 | atpE | - | 100% (86) | F0F1 ATP synthase subunit C |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1337 | atpE | 1e-36 | 90.12% (81) | F0F1 ATP synthase subunit C |
| M. gilvum PYR-GCK | Mflv_2313 | - | 8e-37 | 92.59% (81) | F0F1 ATP synthase subunit C |
| M. tuberculosis H37Rv | Rv1305 | atpE | 1e-36 | 90.12% (81) | F0F1 ATP synthase subunit C |
| M. leprae Br4923 | MLBr_01140 | atpE | 1e-35 | 87.65% (81) | F0F1 ATP synthase subunit C |
| M. abscessus ATCC 19977 | MAB_1448 | - | 3e-37 | 96.05% (76) | F0F1 ATP synthase subunit C |
| M. marinum M | MMAR_4092 | atpE | 1e-36 | 91.36% (81) | ATP synthase C chain AtpE |
| M. avium 104 | MAV_1522 | - | 3e-37 | 90.36% (83) | F0F1 ATP synthase subunit C |
| M. thermoresistible (build 8) | TH_3985 | - | 3e-32 | 80.25% (81) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_3959 | atpE | 9e-37 | 91.36% (81) | F0F1 ATP synthase subunit C |
| M. vanbaalenii PYR-1 | Mvan_4333 | - | 2e-43 | 96.51% (86) | F0F1 ATP synthase subunit C |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01140|M.leprae_Br4923 --MDP--MIAQGALIGGGLIMAGGAIGAGIGDGMAGNALVSGIARQPEAQ
MAV_1522|M.avium_104 MALDP--QVAAAALIGGGLIMGGGAIGAGIGDGIAGNALVSGIARQPEAQ
MSMEG_4941|M.smegmatis_MC2_155 MDLDPNAIITAGALIGGGLIMGGGAIGAGIGDGIAGNALISGIARQPEAQ
Mvan_4333|M.vanbaalenii_PYR-1 MEIDPNAIITAGALIGGGLIMGGGAIGAGIGDGIAGNALIAGIARQPEAQ
MMAR_4092|M.marinum_M --MDP--TIAAGALIGGGLIMAGGAIGAGIGDGIAGNALISGVARQPEAQ
MUL_3959|M.ulcerans_Agy99 --MDP--TIAAGALIGGGLIMAGGAIGAGIGDGIAGNALISGVARQPEAQ
Mb1337|M.bovis_AF2122/97 --MDP--TIAAGALIGGGLIMAGGAIGAGIGDGVAGNALISGVARQPEAQ
Rv1305|M.tuberculosis_H37Rv --MDP--TIAAGALIGGGLIMAGGAIGAGIGDGVAGNALISGVARQPEAQ
MAB_1448|M.abscessus_ATCC_1997 -MADP--TIVAGALIGGGLIMAGGAIGAGIGDGIAGNALISGVARQPEAQ
Mflv_2313|M.gilvum_PYR-GCK --MDP--TIAAGALIGGGLIMAGGAIGAGIGDGIAGNALISGIARQPEAQ
TH_3985|M.thermoresistible__bu --MDP--MIAAGALIGGGLLLGGAATGAAIGDGLAGAALIDGVARQPEAQ
** :. .*******::.*.* **.****:** **: *:*******
MLBr_01140|M.leprae_Br4923 SRLFTPFFITVGLVEAAYFINLAFMALFVFATPVK-
MAV_1522|M.avium_104 GRLFTPFFITVGLVEAAYFINLAFMALFVFATPVGT
MSMEG_4941|M.smegmatis_MC2_155 GRLFTPFFITVGLVEAAYFINLAFMALFVFATPGLQ
Mvan_4333|M.vanbaalenii_PYR-1 GRLFTPFFITVGLVEAAYFINLAFMALFVFATPGLQ
MMAR_4092|M.marinum_M GRLFTPFFITVGLVEAAYFINLAFMALFVFATPVK-
MUL_3959|M.ulcerans_Agy99 GRLFTPFFITVGLVEAAYFINLAFMALFVFATPVK-
Mb1337|M.bovis_AF2122/97 GRLFTPFFITVGLVEAAYFINLAFMALFVFATPVK-
Rv1305|M.tuberculosis_H37Rv GRLFTPFFITVGLVEAAYFINLAFMALFVFATPVK-
MAB_1448|M.abscessus_ATCC_1997 GRLFTPFFITVGLVEAAYFINLAFMALFVFATPGAS
Mflv_2313|M.gilvum_PYR-GCK GRLFTPFFITVGLVEAAYFINLAFMALFVFATPVG-
TH_3985|M.thermoresistible__bu GRLFVPFFITVGLVEAMYFINLAFMALFVFATPVG-
.***.*********** ****************