For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTPAKARPGTREEVDVASVESTEAPWVTIVWDDPVNLMSYVTYVFQKLFGYSEPHATKLMLQVHNEGKAV VSAGSRESMEVDVSKLHAAGLWATMQQDR*
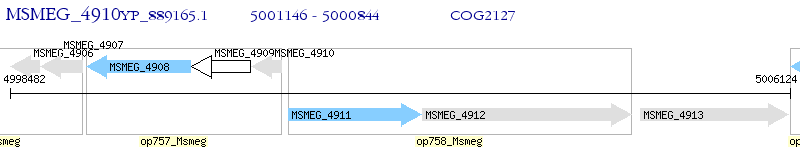
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4910 | clpS | - | 100% (100) | ATP-dependent Clp protease adaptor protein ClpS |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1366 | clpS | 4e-44 | 83.84% (99) | ATP-dependent Clp protease adaptor protein ClpS |
| M. gilvum PYR-GCK | Mflv_2349 | clpS | 1e-46 | 84.00% (100) | ATP-dependent Clp protease adaptor protein ClpS |
| M. tuberculosis H37Rv | Rv1331 | clpS | 4e-44 | 83.84% (99) | ATP-dependent Clp protease adaptor protein ClpS |
| M. leprae Br4923 | MLBr_01165 | clpS | 1e-42 | 84.21% (95) | ATP-dependent Clp protease adaptor protein ClpS |
| M. abscessus ATCC 19977 | MAB_1475 | - | 9e-41 | 75.00% (100) | ATP-dependent Clp protease adaptor protein ClpS |
| M. marinum M | MMAR_4067 | - | 2e-43 | 80.00% (100) | hypothetical protein MMAR_4067 |
| M. avium 104 | MAV_1551 | clpS | 9e-44 | 82.65% (98) | ATP-dependent Clp protease adaptor protein ClpS |
| M. thermoresistible (build 8) | TH_1208 | - | 3e-39 | 87.65% (81) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3931 | - | 5e-36 | 95.71% (70) | hypothetical protein MUL_3931 |
| M. vanbaalenii PYR-1 | Mvan_4296 | clpS | 2e-48 | 87.00% (100) | ATP-dependent Clp protease adaptor protein ClpS |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2349|M.gilvum_PYR-GCK ----MVTPAK----ARPGTREDRDVGEDAAADTPWVTIVWDDPVNLMTYV
Mvan_4296|M.vanbaalenii_PYR-1 ----MVTPAK----ARPGTREDRDVREDATTDAPWVTIVWDDPVNLMTYV
MSMEG_4910|M.smegmatis_MC2_155 ----MVTPAK----ARPGTREEVDVASVESTEAPWVTIVWDDPVNLMSYV
MMAR_4067|M.marinum_M ---MVVASAP----AKPGSVGQQESASRDATASPWVTIVWDDPVNLMTYV
MUL_3931|M.ulcerans_Agy99 --------------------------------------MWDDPVNLMTYV
Mb1366|M.bovis_AF2122/97 ---MAVVSAP----AKPGTTWQRESAPVDVTDRAWVTIVWDDPVNLMSYV
Rv1331|M.tuberculosis_H37Rv ---MAVVSAP----AKPGTTWQRESAPVDVTDRAWVTIVWDDPVNLMSYV
MLBr_01165|M.leprae_Br4923 MWVMVVTLAPIEPKVRPNTTWQHESAPVDITAARWVTIVWDDPVNLMAYV
MAV_1551|M.avium_104 ---MVVMSAPTEPKSRPGTTGQRESAPEDVTASPWVTIVWDDPVNLMTYV
TH_1208|M.thermoresistible__bu ---------------------------VEATDVPWVTIVWDDPVNLMTYV
MAB_1475|M.abscessus_ATCC_1997 ----MGAPAR----ALPGTTAERSVEHVDETQSPWVTIVWDDPVNLMNYV
:******** **
Mflv_2349|M.gilvum_PYR-GCK TYVLQKLFGYTEPHATKLMLQVHNEGKAVVSAGSRESMETDVSRLHAAGL
Mvan_4296|M.vanbaalenii_PYR-1 TYVFQKLFGYSEPHATKLMMQVHNEGKAVVSAGSRESMEVDVTRLHAAGL
MSMEG_4910|M.smegmatis_MC2_155 TYVFQKLFGYSEPHATKLMLQVHNEGKAVVSAGSRESMEVDVSKLHAAGL
MMAR_4067|M.marinum_M TYVFQKLFGYSEPHATKLMLQVHNEGRAVVSAGSREAMEVDVSKLHAAGL
MUL_3931|M.ulcerans_Agy99 TYVFQKLFGYSEPHATKLMLQVHNEGRAVVSAGSREAMEVDVSKLHAAGL
Mb1366|M.bovis_AF2122/97 TYVFQKLFGYSEPHATKLMLQVHNEGKAVVSAGSRESMEVDVSKLHAAGL
Rv1331|M.tuberculosis_H37Rv TYVFQKLFGYSEPHATKLMLQVHNEGKAVVSAGSRESMEVDVSKLHAAGL
MLBr_01165|M.leprae_Br4923 TYVFQKLFGYSEPHATKLMLQVHNEGKAVVSMGSRESMEVDVSKLHAAGL
MAV_1551|M.avium_104 TYVFQKLFGYSEPHATKLMLQVHNEGKAVVSAGSREAMEVDVSKLHAAGL
TH_1208|M.thermoresistible__bu TYVFQKLFGYSEAHATKLMLQVHHEGKAVVSSGSREAMEADVSRLHAAGL
MAB_1475|M.abscessus_ATCC_1997 AYVFQQLFGYSETEATNLMLQVHHEGKAVVSCGARESMEIDVSKLHAAGL
:**:*:****:*..**:**:***:**:**** *:**:** **::******
Mflv_2349|M.gilvum_PYR-GCK WATLQQDR
Mvan_4296|M.vanbaalenii_PYR-1 WATLQQDR
MSMEG_4910|M.smegmatis_MC2_155 WATMQQDR
MMAR_4067|M.marinum_M WATMQQDR
MUL_3931|M.ulcerans_Agy99 WATMQQDR
Mb1366|M.bovis_AF2122/97 WATMQQDR
Rv1331|M.tuberculosis_H37Rv WATMQQDR
MLBr_01165|M.leprae_Br4923 WATMQQDR
MAV_1551|M.avium_104 WATMQQDR
TH_1208|M.thermoresistible__bu WATMQQDR
MAB_1475|M.abscessus_ATCC_1997 WATLQQDR
***:****