For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTEPAAEVSTPRETIRKALVGYRIFAWATGLWLIALCYEMVLKYIVQVDNPPGWIGIVHGWVYFIYLMFT ANLAVKIRWPIGKTIGVLLAGTIPFLGIIVEHFNTRDVKKRFEL
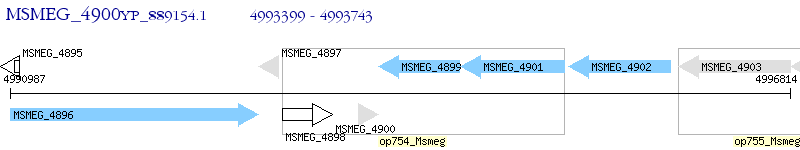
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4900 | - | - | 100% (114) | Pks14 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1377c | - | 8e-46 | 69.30% (114) | hypothetical protein Mb1377c |
| M. gilvum PYR-GCK | Mflv_2362 | - | 1e-42 | 70.87% (103) | hypothetical protein Mflv_2362 |
| M. tuberculosis H37Rv | Rv1342c | - | 8e-46 | 69.30% (114) | hypothetical protein Rv1342c |
| M. leprae Br4923 | MLBr_01176 | - | 3e-36 | 63.73% (102) | hypothetical protein MLBr_01176 |
| M. abscessus ATCC 19977 | MAB_1487c | - | 2e-40 | 66.67% (105) | hypothetical protein MAB_1487c |
| M. marinum M | MMAR_4039 | - | 7e-48 | 73.21% (112) | hypothetical protein MMAR_4039 |
| M. avium 104 | MAV_1559 | - | 1e-37 | 65.45% (110) | hypothetical protein MAV_1559 |
| M. thermoresistible (build 8) | TH_0119 | pks14 | 1e-44 | 72.48% (109) | CONSERVED MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_3905 | - | 6e-48 | 73.21% (112) | hypothetical protein MUL_3905 |
| M. vanbaalenii PYR-1 | Mvan_4284 | - | 2e-44 | 72.38% (105) | hypothetical protein Mvan_4284 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4039|M.marinum_M MTTPETPEAQQTTTTFPVEKIRSALVGYRIMAWTTGIWLIALCYEIVVRY
MUL_3905|M.ulcerans_Agy99 MTPPETPEAQQTTTTFPVEKIRSALVGYRIMAWTTGVWLIALCYEIVVRY
Mb1377c|M.bovis_AF2122/97 MTAPETPAAQHAEPAIAVERIRTALLGYRIMAWTTGLWLIALCYEIVVRY
Rv1342c|M.tuberculosis_H37Rv MTAPETPAAQHAEPAIAVERIRTALLGYRIMAWTTGLWLIALCYEIVVRY
MLBr_01176|M.leprae_Br4923 MTRPETP----QAPDFDFEKSRTALLGYRIMAWTTGIWLIALCYEIVSHL
MAV_1559|M.avium_104 MTTPETPGA--PVPAVPVDKIRTALLGYRIMAWTTGLWLIALCYEIVSHL
Mflv_2362|M.gilvum_PYR-GCK MTAPQSP----SPLT-PVESIRKALTGYRVLAWATGLWLIALCYEMVMKY
Mvan_4284|M.vanbaalenii_PYR-1 MTAPQSP----APIT-PVETIRKALTGYRVLAWATGIWLIALCYEMVMKY
TH_0119|M.thermoresistible__bu MSAPDTP----APITAPIETIRKALLGYRIMAWATGLWLIALCYEMVMKY
MSMEG_4900|M.smegmatis_MC2_155 MTEPAAE------VSTPRETIRKALVGYRIFAWATGLWLIALCYEMVLKY
MAB_1487c|M.abscessus_ATCC_199 MTENETTPDSTVVVTTPVAKIRSALWRYRILAWTTGVWLIALCWEMWLKY
*: : *.** **::**:**:******:*: :
MMAR_4039|M.marinum_M VVKVDNPPTWIGVVHGWVYFAYLLFTANLAVKVRWPIGKTVGVLLSGTIP
MUL_3905|M.ulcerans_Agy99 VVKVDNPPTWIGVVHGWVYFAYLLFTANLAVKVRWPIGKTVGVLLSGTIP
Mb1377c|M.bovis_AF2122/97 VVKVDNPPTWIGVVHGWVYFTYLLLTLNLAVKVRWPLGKTAGVLLAGTIP
Rv1342c|M.tuberculosis_H37Rv VVKVDNPPTWIGVVHGWVYFTYLLLTLNLAVKVRWPLGKTAGVLLAGTIP
MLBr_01176|M.leprae_Br4923 VFHHEIR--WIEVVHGWVYFVYVLTAFNLAIKVRWPIGKTVGVLLAGTVP
MAV_1559|M.avium_104 VMHHEIR--WIEVVHGWVYFVYVLTAFNLAVKVRWPIGKTIGVLLSGTIP
Mflv_2362|M.gilvum_PYR-GCK GFNDDSLG-WIAVVHGWVYFVYLLFTANLAVKVRWPIAKTVGVLLAGTVP
Mvan_4284|M.vanbaalenii_PYR-1 AFQDDSLG-WIAVVHGWVYFVYLLFTANLAVKVRWPIAKTVGILLAGTIP
TH_0119|M.thermoresistible__bu GYGVEGLN-WIAVVHGWVYFVYLLFTANLAVKVRWPIAKTIGVLLAGTIP
MSMEG_4900|M.smegmatis_MC2_155 IVQVDNPPGWIGIVHGWVYFIYLMFTANLAVKIRWPIGKTIGVLLAGTIP
MAB_1487c|M.abscessus_ATCC_199 VEQVPHPPSWIGIVHGWIYFVYLIFTTDLAVKVRWPWPRTIGTLLAGTIP
** :****:** *:: : :**:*:*** :* * **:**:*
MMAR_4039|M.marinum_M LLGIIVEHFQTKDIKTRFAV-----
MUL_3905|M.ulcerans_Agy99 LLGIIVEHFQTKDIKTRFAV-----
Mb1377c|M.bovis_AF2122/97 LLGIVVEHFQTKEIKARFGL-----
Rv1342c|M.tuberculosis_H37Rv LLGIVVEHFQTKEIKARFGL-----
MLBr_01176|M.leprae_Br4923 LLGIIVEHFQTKDVKTRFGLRHSRT
MAV_1559|M.avium_104 LLGIIVEHFQTRDVKARFGV-----
Mflv_2362|M.gilvum_PYR-GCK LVGLIVEHFQTQNIKAQFGI-----
Mvan_4284|M.vanbaalenii_PYR-1 LVGIIVEHFQTQNIKEQFGL-----
TH_0119|M.thermoresistible__bu LVGLIVEHFQTRDLKARFDI-----
MSMEG_4900|M.smegmatis_MC2_155 FLGIIVEHFNTRDVKKRFEL-----
MAB_1487c|M.abscessus_ATCC_199 LLGFIVEHYRTKQVKEQFGI-----
::*::***:.*:::* :* :