For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DFRDSPEEAAFRDRLRSWLADNAASFKASGDDYWARQGEWHQALYGAGFFGTSWPKEFGGQDLPPVYDVI VDEELARAGAPPRPSLGYLVVGLGHHGSKELQQRFLPGMINGTERWCQGFSEPGAGSDLASLITTATRDG DNYIINGHKIWTSYSDVADWCLLLARTDKDVPRHKGISAFIISMHQDGIQQRPLKMINGVTTEFGQVTFD NAVVPASHMVGAPGEGWALAMTVVSHEREPSTLGYSARYGKLVREMASRVEGRAPQDLAWAAVQAEMLRL HVRRRLSEQLDGLKHGPQGSLDKLLMTWVEQSVGHAALAVSGTSDPELLSAYLYSRAQSVMGGTSQIQKN IIASRILGLGV*
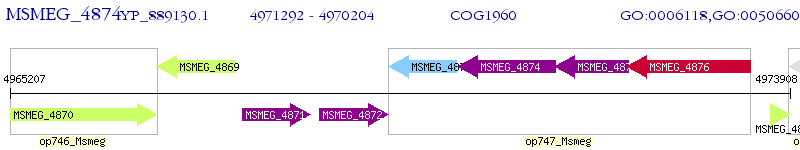
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4874 | - | - | 100% (362) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6012 | - | 4e-58 | 36.53% (386) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4844 | - | 4e-55 | 38.11% (370) | acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3590c | fadE30 | 2e-62 | 37.63% (380) | acyl-CoA dehydrogenase FADE30 |
| M. gilvum PYR-GCK | Mflv_2394 | - | 1e-177 | 81.67% (360) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3560c | fadE30 | 3e-62 | 37.63% (380) | acyl-CoA dehydrogenase FADE30 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 1e-24 | 33.20% (253) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0597 | - | 1e-60 | 37.07% (375) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_4791 | - | 1e-165 | 75.34% (365) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_0865 | - | 1e-171 | 78.33% (360) | acyl-CoA dehydrogenase middle domain-containing protein |
| M. thermoresistible (build 8) | TH_3176 | - | 0.0 | 88.70% (345) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_0360 | - | 1e-131 | 75.68% (296) | acyl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4254 | - | 0.0 | 83.66% (361) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3590c|M.bovis_AF2122/97 --------------MQDV--EEFRAQVRGWLADNLAGEFAALKGLGGPGR
Rv3560c|M.tuberculosis_H37Rv --------------MQDV--EEFRAQVRGWLADNLAGEFAALKGLGGPGR
MAB_0597|M.abscessus_ATCC_1997 MVESQVSPAPAGEPMSEMSDDQVRTEIREWLAENLVGEFAELKGLGGPGS
MSMEG_4874|M.smegmatis_MC2_155 ---------------MDFRDSPEEAAFRDRLRSWLADNAASFKASG----
TH_3176|M.thermoresistible__bu ------------------------------LRNWLADNAAAFTASG----
Mflv_2394|M.gilvum_PYR-GCK ---------------MDFRDSPDEAAFRDRLAAWLKEQKGRFPTSG----
Mvan_4254|M.vanbaalenii_PYR-1 ---------------MDFRDSPEEAAFRDRLRGWLSEQKGKFPTSG----
MMAR_4791|M.marinum_M ---------------MDFRDSPQEAVFRQRLRNWLSVNADDFRGSG----
MUL_0360|M.ulcerans_Agy99 --------------------------------------------------
MAV_0865|M.avium_104 ---------------MDFRDSPDEAAFRERLRSWLSLHAKEFASSG----
MLBr_00737|M.leprae_Br4923 -----MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADV----
Mb3590c|M.bovis_AF2122/97 EHEAFEERRAWNQRLAAAGLTCLGWPEEHGGRGLSTAHRVAFYEEYARAD
Rv3560c|M.tuberculosis_H37Rv EHEAFEERRAWNQRLAAAGLTCLGWPEEHGGRGLSTAHRVAFYEEYARAD
MAB_0597|M.abscessus_ATCC_1997 EHEAFEERLAWNRHLAAAGWTCLGWPVEHGGRGATVSQRVIFYEEYARAE
MSMEG_4874|M.smegmatis_MC2_155 -DDYWARQGEWHQALYGAGFFGTSWPKEFGGQDLPPVYDVIVDEELARAG
TH_3176|M.thermoresistible__bu -DDYWAKQGEWHQALYQAGFFGTSWPKEYGGQDLPPVYDVIVDEELARAG
Mflv_2394|M.gilvum_PYR-GCK -DAYWAKAGEWHQALFEAGFFGTSWPKAYGGQDLPPVYDVIVDEEIAKAG
Mvan_4254|M.vanbaalenii_PYR-1 -DAYWAKAGEWHQALFGAGFFGTSWPKEYGGQDLPPVYDVIVDEEIAKAG
MMAR_4791|M.marinum_M -DQYWARQAHWHQALYREGFFGLSWPREYGGRELAPVYDVIVDEELARAG
MUL_0360|M.ulcerans_Agy99 ---------------------------------------MIVDEELARAG
MAV_0865|M.avium_104 -DDYWARQAEWHQALYREGFFGLSWPREYGGQELAPVYDVIVDEELARAG
MLBr_00737|M.leprae_Br4923 -DQRARFPEEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVD
. ** *:.
Mb3590c|M.bovis_AF2122/97 APDKVNHFGEELLGPTLIAFGTPQQQRRFLPRIRDVTELWCQGYSEPGAG
Rv3560c|M.tuberculosis_H37Rv APDKVNHFGEELLGPTLIAFGTPQQQRRFLPRIRDVTELWCQGYSEPGAG
MAB_0597|M.abscessus_ATCC_1997 APHKVNHLGEELLGPTLIAYGTPEQQQRFLPAIRDVTELWCQGYSEPGAG
MSMEG_4874|M.smegmatis_MC2_155 APPRP-SLGYLVVG--LGHHGSKELQQRFLPGMINGTERWCQGFSEPGAG
TH_3176|M.thermoresistible__bu APPRP-SLGYLVVG--LGHHGSEELQQRFLPGMINGTERWCQGFSEPGAG
Mflv_2394|M.gilvum_PYR-GCK APARP-SLGYLVVG--LSHHGSEELRQRFLPGMINGTERWCQGFSEPGAG
Mvan_4254|M.vanbaalenii_PYR-1 APARP-SLGYLVVG--LSHHGSEELRQRFLPGMINGSERWCQGFSEPGAG
MMAR_4791|M.marinum_M APPRP-SVGYLVFG--IGRHGSDALRQRFLPGIIDGTERWCQGFSEPGAG
MUL_0360|M.ulcerans_Agy99 APPRR-SVGYLVFG--IGRHGSDALRQRFLPGRIDGTERWCQGFSEPGAG
MAV_0865|M.avium_104 APPRP-SVGYLIYG--IGRHASDELRRRFLPGIINGTERWCQGFSEPGAG
MLBr_00737|M.leprae_Br4923 ASASLIPAVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAG
*. : : .: :::.** . . ** **
Mb3590c|M.bovis_AF2122/97 SDLASVATTAELDGDQWVINGQKVWTSLAHLSQWCFVLARTEKGSQRHAG
Rv3560c|M.tuberculosis_H37Rv SDLASVATTAELDGDQWVINGQKVWTSLAHLSQWCFVLARTEKGSQRHAG
MAB_0597|M.abscessus_ATCC_1997 SDLANVSTSAVLDGGDWVINGQKVWTSLALQSQWCFVVARTEPGSSRHKG
MSMEG_4874|M.smegmatis_MC2_155 SDLASLITTATRDGDNYIINGHKIWTSYSDVADWCLLLARTDKDVPRHKG
TH_3176|M.thermoresistible__bu SDLASLTTTATREGDHWVINGHKIWTSYSDVADWCLVLARTDKNVPKHKG
Mflv_2394|M.gilvum_PYR-GCK SDLASLTTTAVKDGDEYVIHGHKIWTSYSDVADWCLVLARTDKDVPKHKG
Mvan_4254|M.vanbaalenii_PYR-1 SDLASLTTTAVRDGDEYVIHGHKIWTSYSDVADWCLLLARTDKDVPKHKG
MMAR_4791|M.marinum_M SDLAALTTTATRVGDDYVVHGHKIWTSYSDVAEWCLLLARTEPGASRHRG
MUL_0360|M.ulcerans_Agy99 SDLAALTTTATRVGDDYVVHGHKIWTSYSDVADWCLLLARTEPGASRHRG
MAV_0865|M.avium_104 SDLASLTTTARLEGDTYVVDGHKIWTSYSDVADWCLLLARTDPQAKRHRG
MLBr_00737|M.leprae_Br4923 SDAASMRTRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPD-KGANG
** * : * * *. :::.* * * : . : * ::* *: *
Mb3590c|M.bovis_AF2122/97 LSYLLVPLDQPGVQIRPIVQITG--TAEFNEVFFDDARTDADLVVGAPGD
Rv3560c|M.tuberculosis_H37Rv LSYLLVPLDQPGVQIRPIVQITG--TAEFNEVFFDDARTDADLVVGAPGD
MAB_0597|M.abscessus_ATCC_1997 LSYLLVPLDQPGVEIRPIIQLTG--TSEFNEVFFDNARTSADLVVGEPGD
MSMEG_4874|M.smegmatis_MC2_155 ISAFIISMHQDGIQQRPLKMINGV-TTEFGQVTFDNAVVPASHMVGAPGE
TH_3176|M.thermoresistible__bu ISAFIVDMHQPGVQQRPLKMINGV-TTEFGQVTFDGAVAPADQMVGEPGE
Mflv_2394|M.gilvum_PYR-GCK ISAFIVSMHQPGIEQRPLKMISGV-TKEFGQVSFDGARVPAENMVGAPGE
Mvan_4254|M.vanbaalenii_PYR-1 ISAFIVNMHQAGIEQRPLKMISGV-TKEFGQVSFDGARVPAENMVGAPGE
MMAR_4791|M.marinum_M LSAFVVAMKQPGVQQRPLRMMNGV-TKEFGQVFFDGATVAADRMVGAPGD
MUL_0360|M.ulcerans_Agy99 LSAFVVAMKQPGVQQRPLRMMNGV-TKEFGQVFFDGATVAADRMIGAPGD
MAV_0865|M.avium_104 LSAFILEMKQPGVQQRPLRMINGV-TNEFGQVFFDGAKVPADRMVGAPGD
MLBr_00737|M.leprae_Br4923 ISAFIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGT
:* ::: .: *.. * * :: ** . .. ::* **
Mb3590c|M.bovis_AF2122/97 GWRVAMATLTFER----------GVSTLGQQIVYARELSNLVELARRTAA
Rv3560c|M.tuberculosis_H37Rv GWRVAMATLTFER----------GVSTLGQQIVYARELSNLVELARRTAA
MAB_0597|M.abscessus_ATCC_1997 GWRVAMGTLTFER----------GVSTLGNQITYARELSRLAELAKSNGA
MSMEG_4874|M.smegmatis_MC2_155 GWALAMTVVSHER----------EPSTLGYSARYGKLVREMASRVEG---
TH_3176|M.thermoresistible__bu GWKLAMTVVSHER----------EPSTLGYSARYGKLVRQMADRVEG---
Mflv_2394|M.gilvum_PYR-GCK GWKLAMTVVSHER----------EPSTLGFSARYGKTVRQMAAGVDG---
Mvan_4254|M.vanbaalenii_PYR-1 GWKLAMTVVSHER----------EPSTLGFSARYGKTVRQMASRLDG---
MMAR_4791|M.marinum_M GWAVAMTVVGHER----------EPSTLGYAARYNKLVAELLARNHAQGH
MUL_0360|M.ulcerans_Agy99 GWAVAMTVVGHER----------EPSTLGYAARYNKLVAELLARNHAQGH
MAV_0865|M.avium_104 GWAVAMTVVGHER----------EPSTLGYAARYGKIVRSLYARTDG---
MLBr_00737|M.leprae_Br4923 GFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESIST-FQ
*: *: .: . * .:*. * : :
Mb3590c|M.bovis_AF2122/97 ADDPLIRERLTRAWTGLRAMRSYALATMEGPAVEQPGQDNVSKLLWANWH
Rv3560c|M.tuberculosis_H37Rv ADDPLIRERLTRAWTGLRAMRSYALATMEGPAVEQPGQDNVSKLLWANWH
MAB_0597|M.abscessus_ATCC_1997 ADDPAIRERLAQAYVGMRTMRAYALATMD---EERPGQDNVSKLLWANWH
MSMEG_4874|M.smegmatis_MC2_155 RAPQDLAWAAVQAEMLRLHVRRRLSEQLDG--LKHGPQGSLDKLLMTWVE
TH_3176|M.thermoresistible__bu RPPEELVWAAVEAEMLRLHVRRRLSEQLDG--IQHGPQGSLDKLLMTWTE
Mflv_2394|M.gilvum_PYR-GCK TPPDELSWAWVQTEMLRLHVRRRLSEQLDG--LTHGPQGSLDKLLMTWVE
Mvan_4254|M.vanbaalenii_PYR-1 APNQELLWAWVQTEMLRLHVRRRLSEQLDG--LTHGPQGSLDKLLMTWVE
MMAR_4791|M.marinum_M PVSDELAWAAVQAEMLTHHVRRRLSEQLDG--VSHGPEGSLDKLLMTWVE
MUL_0360|M.ulcerans_Agy99 PVSEELAWAAVQAEMLTHHVRCRLSEQLDG--VSHGPEGSLDKLLMTWVE
MAV_0865|M.avium_104 PVCEELAWAAVQSEMLTHHVRRRLSEQLDG--VSHGPEGSLDKLLMTWVE
MLBr_00737|M.leprae_Br4923 SIQFMLADMAMKVEAARLIVYAAAARAERG-EPDLGFISAASKCFASDIA
: . : . .* : :
Mb3590c|M.bovis_AF2122/97 RNLGELAMDVIGKPGMTMPDGEFDEWQRLYLFTRADTIYGGSNEIQRNII
Rv3560c|M.tuberculosis_H37Rv RNLGELAMDVIGKPGMTMPDGEFDEWQRLYLFTRADTIYGGSNEIQRNII
MAB_0597|M.abscessus_ATCC_1997 RDLGELAMEIIGTSTLLTNAGEMDEWQRLFLFSRADTIYGGSNEIQRNII
MSMEG_4874|M.smegmatis_MC2_155 QSVGHAALAVSGTS--------DPELLSAYLYSRAQSVMGGTSQIQKNII
TH_3176|M.thermoresistible__bu QSVGHAALAVSGTR--------DPEMLAAYLYSRAQSVMGGTSQIQKNII
Mflv_2394|M.gilvum_PYR-GCK QSVGHAALKTVGTG--------DEELFGAYMYSRAQSVMGGTSQIQKNII
Mvan_4254|M.vanbaalenii_PYR-1 QSVGHAALATVGTS--------DEELFGAYMYSRAQSVMGGTSQIQKNII
MMAR_4791|M.marinum_M QSVGHAALGIAGTH--------DPELFGAYLYSRAQSVMGGTSQIQKNII
MUL_0360|M.ulcerans_Agy99 QSVGHAALGIGGTH--------DPELFGAYLYSRAQSVMGGTSQIQKNII
MAV_0865|M.avium_104 QSVGHAALAVAGTR--------DPDLLSAYLYSRAQSVMGGTSQIQKNII
MLBr_00737|M.leprae_Br4923 MEVTTDAVQLFGGAG----YTSDFPVERFMRDAKITQIYEGTNQIQRVVM
.: *: * :: : *:.:**: ::
Mb3590c|M.bovis_AF2122/97 AERVLGLPREAKG
Rv3560c|M.tuberculosis_H37Rv AERVLGLPREAKG
MAB_0597|M.abscessus_ATCC_1997 SERVLGLPREAR-
MSMEG_4874|M.smegmatis_MC2_155 ASRILGLGV----
TH_3176|M.thermoresistible__bu AGRILGLTMR---
Mflv_2394|M.gilvum_PYR-GCK AQRILGLR-----
Mvan_4254|M.vanbaalenii_PYR-1 AQRILGLGA----
MMAR_4791|M.marinum_M ASRILGLGV----
MUL_0360|M.ulcerans_Agy99 ASRILGLGV----
MAV_0865|M.avium_104 ASRILGL------
MLBr_00737|M.leprae_Br4923 SRALLR-------
: :*