For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MMSSQTFDTVLVANRGEIAVRVIRTLRAMGIRSVAVFSEADAGARHVTEADVAVCIGPAAARHSYLDIDA VVGAARRTDAQAVHPGYGFLSENAEFASALATAGIVFIGPPASAIATMGDKIAAKAAVSAFGVPVVPGIS RPGLADDDLIAGAAEVGYPVLVKPSAGGGGKGMRVVEAAADLPAALVSARREAGAAFGDDTLFLERFVQR PRHIEVQVLADGHGNVIHLGERECSLQRRHQKVIEEAPSPLLDEATRARIGAAACATARSVDYTGAGTVE FIVSADRPDEFFFMEMNTRLQVEHPVTELVTGIDLVEQQIRIAAGEPLAIGQDDITLTGHAVEARVYAED PAAGFLPTGGDVLGLREPTGRGVRVDSGLAAGTVVGSDYDPMLSKIIAHGSDRASALQILDRALADTAVL GVTTNIEFLRFLLADDDVAAGRLDTGLLDRRAPDFAPATVGDEQLIAAAAYLWARQWSAAGGDLWRVPSG WRVGEWAPATFRLHAGDRTDHVYITGNPERASAAVEHGDTHTVSADFTPGSDTFAVTLDGLRTDYRVAVT DSQIWLSGGGRTWSVQKVREEPVRPDDAHSGDAELVSPMPGSVVAVGVPDGSDVTAGTVVVTVEAMKMEH ALTAPVDGVAKILVAVGDQVKVGQPLARITAHTQENES*
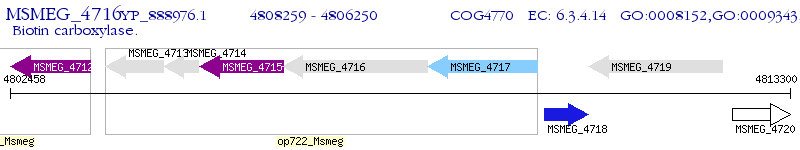
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4716 | - | - | 100% (669) | acetyl-/propionyl-coenzyme A carboxylase alpha chain |
| M. smegmatis MC2 155 | MSMEG_0334 | - | e-152 | 46.24% (679) | acetyl-/propionyl-coenzyme A carboxylase alpha chain |
| M. smegmatis MC2 155 | MSMEG_1807 | - | e-121 | 40.15% (665) | acetyl-/propionyl-coenzyme A carboxylase alpha chain |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2529c | accA1 | 0.0 | 65.14% (657) | acetyl-/propionyl-coenzyme A carboxylase subunit alpha |
| M. gilvum PYR-GCK | Mflv_2553 | - | 0.0 | 77.86% (655) | carbamoyl-phosphate synthase L chain, ATP-binding |
| M. tuberculosis H37Rv | Rv2501c | accA1 | 0.0 | 65.14% (657) | acetyl-/propionyl-coenzyme A carboxylase subunit alpha |
| M. leprae Br4923 | MLBr_00726 | bccA | 1e-116 | 39.12% (662) | acetyl/propionyl CoA carboxylase alpha subunit |
| M. abscessus ATCC 19977 | MAB_4539c | - | 0.0 | 53.75% (666) | putative acyl-CoA carboxylase alpha subunit AccA |
| M. marinum M | MMAR_3848 | accA1 | 0.0 | 67.57% (663) | acetyl-/propionyl-coenzyme A carboxylase alpha chain AccA1 |
| M. avium 104 | MAV_1671 | - | 0.0 | 66.21% (657) | acetyl-/propionyl-coenzyme A carboxylase alpha chain |
| M. thermoresistible (build 8) | TH_3286 | - | 0.0 | 78.51% (656) | PUTATIVE acetyl-/propionyl-coenzyme A carboxylase alpha chain |
| M. ulcerans Agy99 | MUL_3779 | accA1 | 0.0 | 67.12% (663) | acetyl-/propionyl-coenzyme a carboxylase alpha chain AccA1 |
| M. vanbaalenii PYR-1 | Mvan_4090 | - | 0.0 | 77.40% (655) | carbamoyl-phosphate synthase L chain, ATP-binding |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2529c|M.bovis_AF2122/97 -------MFDTVLVANRGEIAVRVIRTLRRLGIRSVAVYSDPDVDARHVL
Rv2501c|M.tuberculosis_H37Rv -------MFDTVLVANRGEIAVRVIRTLRRLGIRSVAVYSDPDVDARHVL
MAV_1671|M.avium_104 -------MFETVLVANRGEIAVRVIRTLRRLGIRSVAVYSDPDAAARHVL
MMAR_3848|M.marinum_M -------MFDTVLVANRGEIAVRVIRTLRRLGIRSVAVYSDADARARHVI
MUL_3779|M.ulcerans_Agy99 -------MFDTVLVANRGEIAVRVIRTLRRLGIRSVAVYSDADARARHVI
MSMEG_4716|M.smegmatis_MC2_155 -MMMSSQTFDTVLVANRGEIAVRVIRTLRAMGIRSVAVFSEADAGARHVT
TH_3286|M.thermoresistible__bu -----------VLVANRGEIAVRVMSTLRRMGIRSVAVFSDADAQARHVA
Mflv_2553|M.gilvum_PYR-GCK -------MFDTVLVANRGEIAVRVIRTLRAMGIRSVAVFSDADAGARHVR
Mvan_4090|M.vanbaalenii_PYR-1 -------MFGTVLVANRGEIAVRVIRTLRAMGIRSVAVFSDADAGARHVA
MAB_4539c|M.abscessus_ATCC_199 ------MTLQSVLIANRGEIAVRIASTLRAMGIRSIAVYTDGDT--DHLG
MLBr_00726|M.leprae_Br4923 MASHASSRIAKVLVANRGEIAVRVIRAARDARLPSVAVYAEPDAEAPHVR
**:*********: : * : *:**::: *. *:
Mb2529c|M.bovis_AF2122/97 EADAAVRLGPAPARESYLDIGKVLDAAARTGAQAIHPGYGFLAENADFAA
Rv2501c|M.tuberculosis_H37Rv EADAAVRLGPAPARESYLDIGKVLDAAARTGAQAIHPGYGFLAENADFAA
MAV_1671|M.avium_104 EADEAVRLGPAAARDSYLNIDKVVETATRTGAQAIHPGYGFLSENADFAA
MMAR_3848|M.marinum_M EADTAVRLGPAAARESYLDIGKVIDAAVRTGAQAIHPGYGFLSENAEFAA
MUL_3779|M.ulcerans_Agy99 EADTAVRLGPAAARESYLDIGKVIDAAVRTGAQAIHPGYGFLSENAEFAA
MSMEG_4716|M.smegmatis_MC2_155 EADVAVCIGPAAARHSYLDIDAVVGAARRTDAQAVHPGYGFLSENAEFAS
TH_3286|M.thermoresistible__bu EADAAVHIGPAAARHSYLDIEAVVRAARRTGAQAVHPGYGFLSENPEFAA
Mflv_2553|M.gilvum_PYR-GCK EADVAINIGPAPARQSYLSIDALLAAITRTGAQAVHPGYGFLSENSQFAD
Mvan_4090|M.vanbaalenii_PYR-1 EADVAVRIGPAPARQSYLSVEALLAAVRRTGAQAVHPGYGFLSENAQFAE
MAB_4539c|M.abscessus_ATCC_199 SCDTAVRLG--ADSSCYLDIDAIISAAKQTGAQAIHPGYGFVSENADFVR
MLBr_00726|M.leprae_Br4923 LADEAFALGGHTSAESYLDFGKILDAAAKSGANAIHPGYGFLAENADFAQ
.* *. :* . .**.. :: : ::.*:*:******::**.:*.
Mb2529c|M.bovis_AF2122/97 ACERARVVFLGPPARAIEVMGDKIAAKNAVAAFDVPVVPGVARAGLTDDA
Rv2501c|M.tuberculosis_H37Rv ACERARVVFLGPPARAIEVMGDKIAAKNAVAAFDVPVVPGVARAGLTDDA
MAV_1671|M.avium_104 ACERAGVVFLGPPARAIQVMGDKITAKNAVAEFDVPVVPGVAEPGLGDDE
MMAR_3848|M.marinum_M ACDRAGVVFLGPPAGAIEVMGDKIAAKNAVSAFDVPVVPGIARPGLSDDD
MUL_3779|M.ulcerans_Agy99 ACDRAGVVFLGSPAGAIEVMGDKIAAKNAVSAFDVPVVPGIARPGLTDDD
MSMEG_4716|M.smegmatis_MC2_155 ALATAGIVFIGPPASAIATMGDKIAAKAAVSAFGVPVVPGISRPGLADDD
TH_3286|M.thermoresistible__bu ALADAGIVFIGPPAAAIRTMGDKIAARAAVSAFGVPVVPGISRPGLTDAD
Mflv_2553|M.gilvum_PYR-GCK ALRSAGVVFIGPPVAAIQTMGDKISAKAAVSEFGVPVVPGISRPGLTDDD
Mvan_4090|M.vanbaalenii_PYR-1 ALDAAGIAFIGPPVAAIQTMGDKISAKAAVSEFGVPVVPGISRPGLTDQD
MAB_4539c|M.abscessus_ATCC_199 ACQAAGIVFVGPPAEAMDAMGDKIRAKGTVAAAGEPVVPGSVG-ALSDAE
MLBr_00726|M.leprae_Br4923 AVIDAGLIWIGPSPQSIRDLGDKVTARHIAARAQAPLVPGTPDPVKNADE
* * : ::*.. :: :***: *: .: *:***
Mb2529c|M.bovis_AF2122/97 LVTAAAEVGYPVLIKPSAGGGGKGMRLVQDPARLPEALVSARREAMSSFG
Rv2501c|M.tuberculosis_H37Rv LVTAAAEVGYPVLIKPSAGGGGKGMRLVQDPARLPEALVSARREAMSSFG
MAV_1671|M.avium_104 LVAAADEIGYPVLIKPSAGGGGKGMRLVEEPGRLREALVGARREAASSFG
MMAR_3848|M.marinum_M LIAAADEVGYPVLVKPSAGGGGKGMRMVEEPARLREALVSARREAASAFG
MUL_3779|M.ulcerans_Agy99 LIAAADEVGYPVLVKPSAGGGGKGMRMVEEPARLREALVSARREAASAFG
MSMEG_4716|M.smegmatis_MC2_155 LIAGAAEVGYPVLVKPSAGGGGKGMRVVEAAADLPAALVSARREAGAAFG
TH_3286|M.thermoresistible__bu LIDGAAQVGFPVLVKPSAGGGGKGMRVVHDPADLPAALAAARREAAAAFG
Mflv_2553|M.gilvum_PYR-GCK LIAGAPDVGFPVLVKPSAGGGGKGMRVVHDAADLAAALASARREAASAFG
Mvan_4090|M.vanbaalenii_PYR-1 LIAGAADVGFPVLVKPSAGGGGKGMRVVQDASELRAALVSARREAASAFG
MAB_4539c|M.abscessus_ATCC_199 LVFTAEKIGAPLIIKPSAGGGGKGMRVVHDLGGISDALVASRREAAALFG
MLBr_00726|M.leprae_Br4923 VVAFAKEHGVPIAIKAAFGGGGKGMKVARTLEEISELYESAVREATVAFG
:: * . * *: :*.: *******::.. : .: *** **
Mb2529c|M.bovis_AF2122/97 DDTLFLERFVLRPRHIEVQVLADAHGNVVHLGERECSLQRRHQKVIEEAP
Rv2501c|M.tuberculosis_H37Rv DDTLFLERFVLRPRHIEVQVLADAHGNVVHLGERECSLQRRHQKVIEEAP
MAV_1671|M.avium_104 DDTLFLERFVLRPRHIEVQILADTHGNVVHLGERECSLQRRHQKVIEEAP
MMAR_3848|M.marinum_M DDTLFLERFVLRPRHIEVQVLADTHGNVVHLGERECSLQRRHQKVIEEAP
MUL_3779|M.ulcerans_Agy99 DDTLFLERFVLRPRHIEVQVLADTHGNVVHLGERECSLQRRHQKVIEEAP
MSMEG_4716|M.smegmatis_MC2_155 DDTLFLERFVQRPRHIEVQVLADGHGNVIHLGERECSLQRRHQKVIEEAP
TH_3286|M.thermoresistible__bu DDTLFLERFVLNPRHIEVQVLADQHGNVIHLGERECSLQRRHQKVIEEAP
Mflv_2553|M.gilvum_PYR-GCK DDTLFLERFVLNPRHIEVQVLADSHGNVVHLGERECSLQRRHQKVIEEAP
Mvan_4090|M.vanbaalenii_PYR-1 DDTLFLERFVLNPRHIEVQVLADAHGNVVHLGERECSLQRRHQKVIEEAP
MAB_4539c|M.abscessus_ATCC_199 DDTLLVERYLARPRHIEVQVFGDTHGNVVHLGERECSLQRRHQKVIEEAP
MLBr_00726|M.leprae_Br4923 RGECFVERYLDKPRHVEAQVIADQHGNIVVAGTRDCSLQRRFQKLVEEAP
. ::**:: .***:*.*::.* ***:: * *:******.**::****
Mb2529c|M.bovis_AF2122/97 SPLLDPQTRERIGVAACNTARCVDYVGAGTVEFIVSAQRPDEFFFMEMNT
Rv2501c|M.tuberculosis_H37Rv SPLLDPQTRERIGVAACNTARCVDYVGAGTVEFIVSAQRPDEFFFMEMNT
MAV_1671|M.avium_104 SPLLDPETRARIGAAACNTARSVDYVGAGTVEFIVSADRPDEFFFMEMNT
MMAR_3848|M.marinum_M SPLLDAPTRARIGAAACNTARSVDYVGAGTVEFIVSGDRPDEFFFMEMNT
MUL_3779|M.ulcerans_Agy99 SPLLDAPTRARIGAAACNTARSVDYVGAGTVELIVSGDRPDEFFFMEMNT
MSMEG_4716|M.smegmatis_MC2_155 SPLLDEATRARIGAAACATARSVDYTGAGTVEFIVSADRPDEFFFMEMNT
TH_3286|M.thermoresistible__bu SPLLDAATRERIGSAACDTARSVDYTGAGTVEFIVSADRPDEFFFMEMNT
Mflv_2553|M.gilvum_PYR-GCK SPLLDPATRARIGAAACDTARSVDYTGAGTVEFIVSADRPDEFFFMEMNT
Mvan_4090|M.vanbaalenii_PYR-1 SPLLDPQTRARIGAAACDTARSVDYTGAGTVEFIVSADRPDEFFFMEMNT
MAB_4539c|M.abscessus_ATCC_199 SALLTPQLRAAMGQTACQIARSVGYFGAGTVEFILDSDAPESYYFMEMNT
MLBr_00726|M.leprae_Br4923 APFLTDAQRKEIHESAKRICKEAHYYGAGTVEYLVGQD--GLISFLEVNT
:.:* * : :* .: . * ****** ::. : *:*:**
Mb2529c|M.bovis_AF2122/97 RLQVEHPVTEAITGLDLVEWQLRVGAGEKLGFAQNDIELRGHAIEARVYA
Rv2501c|M.tuberculosis_H37Rv RLQVEHPVTEAITGLDLVEWQLRVGAGEKLGFAQNDIELRGHAIEARVYA
MAV_1671|M.avium_104 RLQVEHPVTEAVTGLDLVEWQLRVAAGEKLVFAQDDIELRGHAIEARVYA
MMAR_3848|M.marinum_M RLQVEHPVTEAVTGLDLVEWQLRVAAGEKLGFAQDDIELNGHAIEARVYA
MUL_3779|M.ulcerans_Agy99 RLQVEHPVTEAVTGLDLVEWQLRVAAGEKLGFAQDDIELNGHAIEARVYA
MSMEG_4716|M.smegmatis_MC2_155 RLQVEHPVTELVTGIDLVEQQIRIAAGEPLAIGQDDITLTGHAVEARVYA
TH_3286|M.thermoresistible__bu RLQVEHPVTEMVTGVDLVEQQIRIAAGEKLAIAQDDVTLSGHAVEARIYA
Mflv_2553|M.gilvum_PYR-GCK RLQVEHPVTEMVTGLDLVELQVRIAAGETLPIAQDDIHLDGHAIEARVYA
Mvan_4090|M.vanbaalenii_PYR-1 RLQVEHPVTEMVTGLDLVELQVRIAAGEKLPIAQDDIRLDGHAVEARVYA
MAB_4539c|M.abscessus_ATCC_199 RLQVEHPVTEMVTGIDLVQWQILVAAGERLPLSQEQIVLTGHAVEARVYA
MLBr_00726|M.leprae_Br4923 RLQVEHPVTEETTGIDLVLQQFKIANGEKLELIKDPIPC-GHAIEFRING
********** **:*** *. :. ** * : :: : ***:* *: .
Mb2529c|M.bovis_AF2122/97 EDPAREFLPTGGRVLAVFEPAGPGVRVDSSLLGGTVVGSDYDPLLTKVIA
Rv2501c|M.tuberculosis_H37Rv EDPAREFLPTGGRVLAVFEPAGPGVRVDSSLLGGTVVGSDYDPLLTKVIA
MAV_1671|M.avium_104 EDPARGFLPTGGRVLGVFEPSGDGVRVDSSLLPGTVVGSDYDPMLSKVIA
MMAR_3848|M.marinum_M EDPARGFLPTGGRVLQVAEPSGTGIRVDSSLLADTVVGSDYDPMLSKVIA
MUL_3779|M.ulcerans_Agy99 EDPARGFLPTGGRVLQVAEPSGTGIRVDSSLLADTVVGSDYDPMVSKVIA
MSMEG_4716|M.smegmatis_MC2_155 EDPAAGFLPTGGDVLGLREPTGRGVRVDSGLAAGTVVGSDYDPMLSKIIA
TH_3286|M.thermoresistible__bu EDPAAGFLPTGGTVLHVAEPDHPGVRVDSGLFDGTVVGSDYDPMLAKVIA
Mflv_2553|M.gilvum_PYR-GCK EDPARGFLPTGGHVLGLREPAGPGVRVDSGLVRGTVVGSDYDPMLAKVIT
Mvan_4090|M.vanbaalenii_PYR-1 EDPARGFLPTGGDVLALREPAGAGVRVDSGLTRGTAVGSDYDPMLAKVIV
MAB_4539c|M.abscessus_ATCC_199 EDPAREFLPTGG-TVALARFSG-TARTDTALATGSVVGSRFDPMLAKVIV
MLBr_00726|M.leprae_Br4923 EDAGRNFLPSPGPVSKFHPPTGPGVRLDSGVETGSVIGGQFDSMLAKLIV
**.. ***: * . . * *:.: .:.:*. :*.:::*:*.
Mb2529c|M.bovis_AF2122/97 HGADREEALDRLDQALARTAVLGVQTNVEFLRFLLADERVRVGDLDTAVL
Rv2501c|M.tuberculosis_H37Rv HGADREEALDRLDQALARTAVLGVQTNVEFLRFLLADERVRVGDLDTAVL
MAV_1671|M.avium_104 HGADRDEALARLDRALSHTAILGVQTNVEFLRFLLADERVRAGDLDTALL
MMAR_3848|M.marinum_M HGANREQALERLDRALAHTAILGVQTNVEFLRFLLADERVRAGDLDTALL
MUL_3779|M.ulcerans_Agy99 HGANREQALERLDRALAHTAILGVQTNVEFLRFLLADERVRAGDLDTALL
MSMEG_4716|M.smegmatis_MC2_155 HGSDRASALQILDRALADTAVLGVTTNIEFLRFLLADDDVAAGRLDTGLL
TH_3286|M.thermoresistible__bu HGPDRATALRTLDRALADTAILGLTTNIEFLRFLLADDDVAAGRLDTGLL
Mflv_2553|M.gilvum_PYR-GCK HAQDRPAALRALDAALADTAVLGLTTNVEFLRFLLADPDVAAGDLDTGLL
Mvan_4090|M.vanbaalenii_PYR-1 HAGDRPTALRALDAALADTAVLGLTTNVEFLRFLLADPDVAAGNLDTGLL
MAB_4539c|M.abscessus_ATCC_199 HAADRDTALAEVDRALAETRILGVGTNIDFLRALVTNPVVMSGDIDTALL
MLBr_00726|M.leprae_Br4923 HGATRQEALARARRALDEFEVEGLATVIPFHRAVVSDPALIGDNNSFSVH
*. * ** ** : *: * : * * :::: : . . .:
Mb2529c|M.bovis_AF2122/97 DERSADFTARPAPDDVLAAGGLYRQWALARRAQ---GDLWAAPSGWRGGG
Rv2501c|M.tuberculosis_H37Rv DERSADFTARPAPDDVLAAGGLYRQWALARRAQ---GDLWAAPSGWRGGG
MAV_1671|M.avium_104 DARLADFTPLPAPDDVLAAAGLYRQSALAARARRFAGNPWAAPTGWRAGG
MMAR_3848|M.marinum_M DERLADFVPLAAPDDVFAAAGLYVQQALSRRAA---GSLWAAPSGWRVGG
MUL_3779|M.ulcerans_Agy99 DERLADFVPLAAPDDVFAAAGLYVQQALSRRAA---GSLWAAPSGWRVGG
MSMEG_4716|M.smegmatis_MC2_155 DRRAPDFAPATVGDEQLIAAAAYLWARQWSAAG---GDLWRVPSGWRVGE
TH_3286|M.thermoresistible__bu DRCAPDFTPAGATDAELIAAGAYHWLQRWADAA---DDLWSQPSGWRVGA
Mflv_2553|M.gilvum_PYR-GCK DRRLPDFAPAPPTDDDLIAGAAYRWLRSWPTAP---ADPWDVPSGWRIAG
Mvan_4090|M.vanbaalenii_PYR-1 DRRLPDFAPAPATDDELIAAAAYRWLRSWPVAP---SSPWEVPSGWRIAG
MAB_4539c|M.abscessus_ATCC_199 DKIAAEYHPPAAPPWVWVAASALLDGQSG--AG----TLWDGKSGWRVGA
MLBr_00726|M.leprae_Br4923 TR------------------WIETEWN-----------------------
Mb2529c|M.bovis_AF2122/97 HMAPVRTAMRTPLRSETVSVWGPPESAQVQVGDGEIDCASVQVT--REQM
Rv2501c|M.tuberculosis_H37Rv HMAPVRTAMRTPLRSETVSVWGPPESAQVQVGDGEIDCASVQVT--REQM
MAV_1671|M.avium_104 SPAPVRTDMRTALRTETVSVWGPPEAAEVQVGAGERLAASAHLG--PDRI
MMAR_3848|M.marinum_M VTAPVRTAMQTPLRTETVSVFGLPEGAHVQVGDGEMLSASVQLA--PGVM
MUL_3779|M.ulcerans_Agy99 VTAPVRTAMQTPLRTETVSVFGLPEGAHVQVGDGEMLLASVQLA--PGVM
MSMEG_4716|M.smegmatis_MC2_155 W-APATFRLHAGDRTDHVYITGNPERASAAVEHGDTHTVSADFTPGSDTF
TH_3286|M.thermoresistible__bu Q-AATSYRLEAGDRTDHVRLTGHPYDATVAIEDGDPRPLTAALT-GS-RL
Mflv_2553|M.gilvum_PYR-GCK R-APTSIRLHAGERTDHVWLTGAPGDATATVEGGETRTLRASLD--GDLL
Mvan_4090|M.vanbaalenii_PYR-1 R-AATPFRLHAGERTDHVWLTGTPGDATATVEGGRTRTLRASLV--ADRL
MAB_4539c|M.abscessus_ATCC_199 H-APRAHRLRIGDSTELVRLWGDP-VTEVSIGEGGRTPSRVHCA--TAEV
MLBr_00726|M.leprae_Br4923 -------------------NTIEPFIDNQPLDEEDTRPQ--------QTV
* : .
Mb2529c|M.bovis_AF2122/97 SVTISGLRRDYRWAEADRHLWIADERGTWHLREAEEHKIH--RAVGAR-P
Rv2501c|M.tuberculosis_H37Rv SVTISGLRRDYRWAEADRHLWIADERGTWHLREAEEHKIH--RAVGAR-P
MAV_1671|M.avium_104 SVTLDGRRRDYRWAEADRHLWIADERGTWHLREAEQTTIH--RAAGPQ-R
MMAR_3848|M.marinum_M SVTVDGLRRDYRWAEAERHLWIADVRGTWHIREAEEHKIH--RAAGER-P
MUL_3779|M.ulcerans_Agy99 SVTVDGLRRDYRWAEAERHLWIADVRGTWHIREAEEHKIH--RAAGER-P
MSMEG_4716|M.smegmatis_MC2_155 AVTLDGLRTDYRVAVTDSQIWLSGGGRTWSVQKVREEPVR--PDDAHSGD
TH_3286|M.thermoresistible__bu TVTVDGLRTDYTVAAADGRLWLAAAGRAVAVSEVREAPVR--PDDEHRGD
Mflv_2553|M.gilvum_PYR-GCK AVTIDGLRTEYLVAEAPGQVWLAGAGRVTILEEVREAPVR--PDDEHSGD
Mvan_4090|M.vanbaalenii_PYR-1 AVTVDGMRAEYLVAGAGGQVWLAGPDRVTMVEEVREAPVR--PDDAHSGD
MAB_4539c|M.abscessus_ATCC_199 SVEFAGQLHRADAVRSDAAAWVATGDGTWHADIAPEPRLR--HLDGDDED
MLBr_00726|M.leprae_Br4923 IVEVDGRRLEVSLPADLALANPAGCNPAGVIRKKPKPRKRGGHTGAATSG
* . * : . : :
Mb2529c|M.bovis_AF2122/97 AEVVSPMPGSVIAVQVESGSQISAGDVVVVVEAMKMEHSLEAPVSGRVQ-
Rv2501c|M.tuberculosis_H37Rv AEVVSPMPGSVIAVQVESGSQISAGDVVVVVEAMKMEHSLEAPVSGRVQ-
MAV_1671|M.avium_104 AEILSPMPGSVIAVQTPSGSDVSEGDVVVVVEAMKMEHSLAAPVSGRVE-
MMAR_3848|M.marinum_M AEVVSPMPGNVIAVQAESGTAVSEGDVVVIIEAMKMEHPLVAPISGRVE-
MUL_3779|M.ulcerans_Agy99 AEVVSPMPGNVIAVRAESGTAVSEGDVVVIIEAMKMEHPLVAPISGRVE-
MSMEG_4716|M.smegmatis_MC2_155 AELVSPMPGSVVAVGVPDGSDVTAGTVVVTVEAMKMEHALTAPVDGVAK-
TH_3286|M.thermoresistible__bu AELVSPMPGSVVALGVSDGDAVDAGTLVVTVEAMKMEHPLTAPVAGVVE-
Mflv_2553|M.gilvum_PYR-GCK AELTSPMPGTVVAVGVEDGAAVTAGTVVVTVEAMKMEHALSAPVDGVVE-
Mvan_4090|M.vanbaalenii_PYR-1 AELTSPMPGAVVAVGVDDGATVTAGTVVVTVEAMKMEHALAAPVDGVVE-
MAB_4539c|M.abscessus_ATCC_199 GEIRSSMPGVVRVVSVSTGAAVEREAAVVVVEAMKMEHTLRAPIAGTAT-
MLBr_00726|M.leprae_Br4923 DAVTAPMQGTVVKVAVAEGQTVMTGDLVVVLEAMKMENPVTAHKDGIITG
: :.* * * : . * : ** :******:.: * *
Mb2529c|M.bovis_AF2122/97 VLVSVGDQVKVEQVLARIKD---------------
Rv2501c|M.tuberculosis_H37Rv VLVSVGDQVKVEQVLARIKD---------------
MAV_1671|M.avium_104 VLVCVGDQVSVDQVLARVVPEESDPPADSSKDET-
MMAR_3848|M.marinum_M VLVAVGDQVKVEQVLARLIPDTEQAQSKDSAESKD
MUL_3779|M.ulcerans_Agy99 VLVAVGDQVKVEQVLARLIPDTEQAQSKDSAESKD
MSMEG_4716|M.smegmatis_MC2_155 ILVAVGDQVKVGQPLARITAHTQENES--------
TH_3286|M.thermoresistible__bu LLVAVGDQVKVGQPLARITTEQED-----------
Mflv_2553|M.gilvum_PYR-GCK LLVDAGEQVKVGQVLARITASEEPKA---------
Mvan_4090|M.vanbaalenii_PYR-1 LLVSAGEQVKVGQVLARITAIEEPQS---------
MAB_4539c|M.abscessus_ATCC_199 VSVAVGDQVAVDQLLATIHPNTNPEEGKP------
MLBr_00726|M.leprae_Br4923 LAVEAGTAITQGTVLAEIK----------------
: * .* : ** :