For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLYRDRAVVLRQHKLGEADRIVTLLTRDHGLVRAVAKGVRRTRSKFGARLEPFAHIDVQLHPGRNLDIVT QVQAIDAFAADIVSDYGRYTTACAMLETAERLAGEERAPMPDLHRLTVAALRAIADGRRARELVLDSYLL RAMAIAGWAPALTECARCAAPGPHRAFHVAAGGSVCVHCRPSGSSTPPQAVLELMSALHDGDWEYAESST PPHRSQASGLIAAHLQWHLERQLRTLPLVERGYRVDRTVAEQRAALVRQDMAHGNHTGQEDLPATASGA*
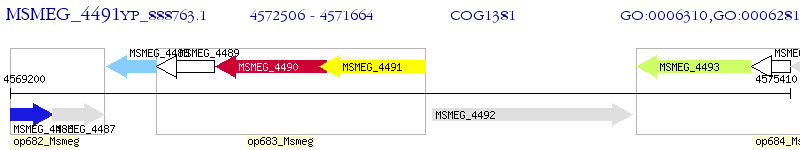
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4491 | recO | - | 100% (280) | DNA repair protein RecO |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2383c | recO | 1e-126 | 81.13% (265) | DNA repair protein RecO |
| M. gilvum PYR-GCK | Mflv_2713 | recO | 1e-133 | 83.93% (280) | DNA repair protein RecO |
| M. tuberculosis H37Rv | Rv2362c | recO | 1e-126 | 81.13% (265) | DNA repair protein RecO |
| M. leprae Br4923 | MLBr_00633 | recO | 1e-127 | 82.46% (268) | DNA repair protein RecO |
| M. abscessus ATCC 19977 | MAB_1675 | recO | 1e-122 | 82.81% (256) | DNA repair protein RecO |
| M. marinum M | MMAR_3672 | recO | 1e-127 | 82.64% (265) | recombination protein O RecO |
| M. avium 104 | MAV_2033 | recO | 1e-124 | 82.81% (256) | DNA repair protein RecO |
| M. thermoresistible (build 8) | TH_0840 | - | 1e-137 | 85.61% (278) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3615 | recO | 1e-127 | 82.26% (265) | DNA repair protein RecO |
| M. vanbaalenii PYR-1 | Mvan_3823 | recO | 1e-134 | 83.63% (281) | DNA repair protein RecO |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2713|M.gilvum_PYR-GCK -------MRLYRDRAVVLRQHKLGEADRIVSLLTREHGLVRAVAKGVRRT
Mvan_3823|M.vanbaalenii_PYR-1 MQWETGPMRLYRDRAVVLRQHKLGEADRIVSLLTRDHGLVRAVAKGVRRT
MSMEG_4491|M.smegmatis_MC2_155 -------MRLYRDRAVVLRQHKLGEADRIVTLLTRDHGLVRAVAKGVRRT
MAB_1675|M.abscessus_ATCC_1997 -------MRLYRDRAVVLRQHKLGEADRIVTLLTRQHGLVRAVAKGVRRT
TH_0840|M.thermoresistible__bu -------MRLYRDRAVVLRQHKLGEADRIVTLLTREHGLVRAVAKGVRRT
Mb2383c|M.bovis_AF2122/97 -------MRLYRDRAVVLRQHKLGEADRIVTLLTRDHGLVRAVAKGVRRT
Rv2362c|M.tuberculosis_H37Rv -------MRLYRDRAVVLRQHKLGEADRIVTLLTRDHGLVRAVAKGVRRT
MMAR_3672|M.marinum_M -------MRLYRDRAVVLRQHKLGEADRIVTLLTRDHGLVRAVAKGVRRT
MUL_3615|M.ulcerans_Agy99 -------MRLYRDRAVVLRQHKLGEADRIVTLLTRDHGLVRAVAKGVRRT
MLBr_00633|M.leprae_Br4923 -------MRLYRDWALVLRQHNLGEADRIVTLLTRDHGLVRAVAKGVRRT
MAV_2033|M.avium_104 ----------------MLRQHKLGEADRIVTLLTRDHGLVRAVAKGVRRT
:****:********:****:**************
Mflv_2713|M.gilvum_PYR-GCK RSKFGARLEPFAHIDVQLHPGRNLDIVTQVQAIDAFASDIVSDYGRYTCA
Mvan_3823|M.vanbaalenii_PYR-1 RSKFGARLEPFAHIDVQLHPGRNLDIVTQVQAIDAFASDIVSDYGRYTCA
MSMEG_4491|M.smegmatis_MC2_155 RSKFGARLEPFAHIDVQLHPGRNLDIVTQVQAIDAFAADIVSDYGRYTTA
MAB_1675|M.abscessus_ATCC_1997 RSKFGSRLEPFAHIDVQLHPGRNLDIVTQVQAIDAFAADIVSDYGRYTTA
TH_0840|M.thermoresistible__bu RSKFGARLEPFAHIDVQLHPGRNLDIVTQVQAIDAFAADIVSDYGRYTCA
Mb2383c|M.bovis_AF2122/97 RSKFGARLEPFAHIEVQLHPGRNLDIVTQVVSVDAFATDIVADYGRYTCG
Rv2362c|M.tuberculosis_H37Rv RSKFGARLEPFAHIEVQLHPGRNLDIVTQVVSVDAFATDIVADYGRYTCG
MMAR_3672|M.marinum_M RSKFGSRLEPFAHIDAQLHPGRNLDIVTQVVSIDAFAADIVNDYGRYTCG
MUL_3615|M.ulcerans_Agy99 RSKFGSRLEPFAHIDAQLHPGRNLDIVTQVVSIDAFAADIVNDYGRYTCG
MLBr_00633|M.leprae_Br4923 RSKFGARLEPFAYIDAQLHPGRNLDIVTQVVSIDAFATDIVSDYGRYTCG
MAV_2033|M.avium_104 RSKFGARLEPFAHIDAQLHPGRNLDIVTQVVSIDAFATDIVSDYGRYTCA
*****:******:*:.************** ::****:*** ****** .
Mflv_2713|M.gilvum_PYR-GCK CAMLETAERIAGEERAPVPALHRLTVAALRAVADGTRPRELVLDAYLLRA
Mvan_3823|M.vanbaalenii_PYR-1 CAMLETAERIAGEERAPVPALHKLTVAALRAVADGTRPRELVLDAYLLRA
MSMEG_4491|M.smegmatis_MC2_155 CAMLETAERLAGEERAPMPDLHRLTVAALRAIADGRRARELVLDSYLLRA
MAB_1675|M.abscessus_ATCC_1997 CAILETAERIAGEERAPAVALHRLTVGALRAIADQQRSRELILDAYLLRA
TH_0840|M.thermoresistible__bu CAMLETAERLAGEERAPVPALHRLTVAALRAVADGSRPRDLVLDAYLLRA
Mb2383c|M.bovis_AF2122/97 CAILETAERLAGEERAPAPALHRLTVGALRAVADGQRPRDLLLDAYLLRA
Rv2362c|M.tuberculosis_H37Rv CAILETAERLAGEERAPAPALHRLTVGALRAVADGQRPRDLLLDAYLLRA
MMAR_3672|M.marinum_M CAMLETAERLAGEERAPAPALHRLTVGALRAVADGSRPRDLLLDAYLLRA
MUL_3615|M.ulcerans_Agy99 CAMLETAERLAGEERAPAPALHRLTVGALRAVADGSRPRDLLLDAYLLRA
MLBr_00633|M.leprae_Br4923 CAMLETAERLAGEERAPAPTLHRLTVSALRAVADGNRPRDLLLDAYLLRA
MAV_2033|M.avium_104 CAMLETAERLAGEERAPAPALHGLTVSALRAVADGRRSRDLLLDAYLLRA
**:******:******* ** ***.****:** *.*:*:**:*****
Mflv_2713|M.gilvum_PYR-GCK MGVAGWAPALIECARCAAPGPHRAFHVAAGGSVCVHCRPSGSVTPPQGVL
Mvan_3823|M.vanbaalenii_PYR-1 MGVAGWAPSLIECARCAAPGPHRAFHVAAGGSVCVHCRPSGSVTPPQAVL
MSMEG_4491|M.smegmatis_MC2_155 MAIAGWAPALTECARCAAPGPHRAFHVAAGGSVCVHCRPSGSSTPPQAVL
MAB_1675|M.abscessus_ATCC_1997 MSIAGWAPAIGECARCATPGPHRAFHVAAGGSVCVHCRPAGAVTPPQGVL
TH_0840|M.thermoresistible__bu MGIAGWAPALTECARCAAPGPHRAFHIAAGGSVCLHCRPAGAVTPPQAVL
Mb2383c|M.bovis_AF2122/97 MGIAGWAPALTECARCATPGPHRAFHIATGGSVCAHCRPAGSTTPPLGVV
Rv2362c|M.tuberculosis_H37Rv MGIAGWAPALTECARCATPGPHRAFHIATGGSVCAHCRPAGSTTPPLGVV
MMAR_3672|M.marinum_M MGIAGWAPALTECARCATPGPHRAFHIAAGGSVCPHCRPAGSTTPPLGVL
MUL_3615|M.ulcerans_Agy99 MGIAGWVPALTECARCATPGPHRAFHIAAGGSVCPHCRPAGSTTPPLGVL
MLBr_00633|M.leprae_Br4923 MGIAGWAPALTACARCATPGPHRAFHIAAGGSVCVHCRPAGSTTPPRGVL
MAV_2033|M.avium_104 MGIAGWAPALTECARCATPGPHRAFHVGAGGSVCPHCRPAGSTTPPPGVL
*.:***.*:: *****:********:.:***** ****:*: *** .*:
Mflv_2713|M.gilvum_PYR-GCK DLMAALYDGDWEHAQISTPAHRSQASGLVAAHLQWHLERQLRTLPLVERP
Mvan_3823|M.vanbaalenii_PYR-1 DLMAALYEGDWEHAQISTTAHRSQASGLVAAHLQWHLERQLRTLPLVERV
MSMEG_4491|M.smegmatis_MC2_155 ELMSALHDGDWEYAESSTPPHRSQASGLIAAHLQWHLERQLRTLPLVERG
MAB_1675|M.abscessus_ATCC_1997 ELMAALHDGDWAATDDVPQTQRTQASGLIAAHLQWHLERKLRTLPLVERG
TH_0840|M.thermoresistible__bu ELMSALHDGDWEFAQASSPAHRSQASGLIAAHLQWHLERKLRTLPLVERS
Mb2383c|M.bovis_AF2122/97 DLMSALYDGDWEAAEAAPQSARSHVSGLVAAHLQWHLERQLKTLPLVERF
Rv2362c|M.tuberculosis_H37Rv DLMSALYDGDWEAAEAAPQSARSHVSGLVAAHLQWHLERQLKTLPLVERF
MMAR_3672|M.marinum_M DLMSALHDGDWEAAQGAPQSHRSYVSGLVAAHLQWHLERQLKTLPLVERF
MUL_3615|M.ulcerans_Agy99 DLMSALHDGDWEAAQGAPQSHRSYVSGLVAAHLQWHLERQLKTLPLVERF
MLBr_00633|M.leprae_Br4923 ELMSALHDGDWGTAQQAPQSHRSYASGLVAAHLQWHLERQLKTLPLVERT
MAV_2033|M.avium_104 DLMSALHDGDWEFAEQTPQSHRNYVSGLVAAHLQWHLERQLKTLPLVERT
:**:**::*** :: . . *. .***:**********:*:*******
Mflv_2713|M.gilvum_PYR-GCK G---RAE--VARN------------IRQDMAHGNQA--EEQLPPAASGA
Mvan_3823|M.vanbaalenii_PYR-1 Y---RAERMAASG------------IRQDVAHGNQADGEEGLPAAAPGA
MSMEG_4491|M.smegmatis_MC2_155 Y---RVDRTVAEQRAAL--------VRQDMAHGNHT-GQEDLPATASGA
MAB_1675|M.abscessus_ATCC_1997 -------------------------SRLNMSHG-----EEQPARSAAG-
TH_0840|M.thermoresistible__bu H---RVDRTLAEHRAAVAADHRAAGFGQDMAHGVEA-GRGPDGQAVVG-
Mb2383c|M.bovis_AF2122/97 ---YQADRSVAERRAAL--------IGQDIAGG----------------
Rv2362c|M.tuberculosis_H37Rv ---YQADRSVAERRAAL--------IGQDIAGG----------------
MMAR_3672|M.marinum_M ---YQADRSVAERRAAL--------IGQDSECG----------------
MUL_3615|M.ulcerans_Agy99 ---YQADRSVAERRAAL--------IGQDSECG----------------
MLBr_00633|M.leprae_Br4923 HQAYQVDRSIAERRAAL--------VRQDMACG----------------
MAV_2033|M.avium_104 ---YHIDRTIADQRATL--------IGQDMDCG----------------
: *