For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAAMATYLIDLSPEDMQRRLPDALRVYVDAMRYPRGTEEQRAALWLEHTRRQGWKAVAAVEFGTDDGSHP ETPSRDDLVAAPLLGIAYGYCGAPDQWWQQQVVSGLHRIGATREQIAELMSSYFELTELHIEPRAQGRGL GEALVRRLLADRGESHVLLSTPEINGEANRAWRLYRRLGFTDVIRGYHFAGDPRAFAILGRPLPL
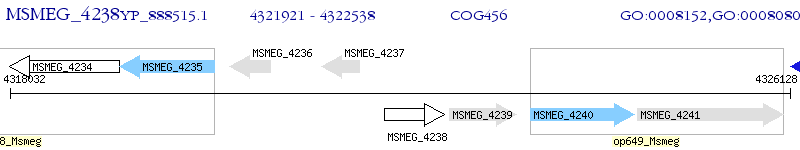
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4238 | - | - | 100% (205) | hypothetical protein MSMEG_4238 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2192 | - | 3e-80 | 72.33% (206) | hypothetical protein Mb2192 |
| M. gilvum PYR-GCK | Mflv_2977 | - | 9e-89 | 78.71% (202) | GCN5-related N-acetyltransferase |
| M. tuberculosis H37Rv | Rv2170 | - | 3e-80 | 72.33% (206) | hypothetical protein Rv2170 |
| M. leprae Br4923 | MLBr_00903 | - | 8e-77 | 66.67% (210) | hypothetical protein MLBr_00903 |
| M. abscessus ATCC 19977 | MAB_1995c | - | 1e-69 | 64.04% (203) | hypothetical protein MAB_1995c |
| M. marinum M | MMAR_3205 | - | 1e-77 | 67.61% (213) | hypothetical protein MMAR_3205 |
| M. avium 104 | MAV_2324 | - | 4e-81 | 71.78% (202) | acetyltransferase, gnat family protein |
| M. thermoresistible (build 8) | TH_0706 | - | 2e-91 | 80.20% (202) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3513 | - | 8e-78 | 67.61% (213) | hypothetical protein MUL_3513 |
| M. vanbaalenii PYR-1 | Mvan_3534 | - | 5e-84 | 74.38% (203) | GCN5-related N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4238|M.smegmatis_MC2_155 MAAMATYLIDLSPEDMQRRLPDALRVYVDAMRYPRGTEEQRAALWLEHTR
TH_0706|M.thermoresistible__bu ---LATYLIDLSPSDMQRRLRDALTVYVDAMRYPRGTENQRAGMWLEHTR
Mflv_2977|M.gilvum_PYR-GCK ---MATRLIDLSPSDMQQRLGDALAVYVDAMRYPRGTEDQRASMWLEHIR
Mvan_3534|M.vanbaalenii_PYR-1 ---MAARLIDLSPGDMQQRLREALAVYVDAMHYPRGTEDQRASMWLEHIR
Mb2192|M.bovis_AF2122/97 ---MAIFLIDLPPSDMERRLGDALTVYVDAMRYPRGTETLRAPMWLEHIR
Rv2170|M.tuberculosis_H37Rv ---MAIFLIDLPPSDMERRLGDALTVYVDAMRYPRGTETLRAPMWLEHIR
MAV_2324|M.avium_104 ---MAIFLIDLLPHDMERRLGDALAVYVDAMRYPRGTENQRAAMWLEHIR
MLBr_00903|M.leprae_Br4923 ---MAIFLINLSPNEMERRLNEALEVYVDAMRYPRNTENLRAGIWLEHIR
MMAR_3205|M.marinum_M ---MTIFVIDLPPDEMERRLREALTVYVDAMRYPRGTENQRAAMWLEHVR
MUL_3513|M.ulcerans_Agy99 ---MTIFVIDLPPDEMERRLREALTVYVDAMRYPRGTENQRAAMWLEHVR
MAB_1995c|M.abscessus_ATCC_199 ---MTTFLADLSQRDMQRRLSEALRVYVIAMGYPHGTEDQRAPMWLEHSR
:: : :* :*::** :** *** ** **:.** ** :**** *
MSMEG_4238|M.smegmatis_MC2_155 RQGWKAVAAVEFG--------TDDGSH--PETPSRDDLVAAPLLGIAYGY
TH_0706|M.thermoresistible__bu RPGWKAVAAVEV---------PDGAPG--DSPPSQATLADAPLLGIAYGY
Mflv_2977|M.gilvum_PYR-GCK RHGWKAVAAVEVS--------GDPTDA--DHLPAR-----APLLGIAYGY
Mvan_3534|M.vanbaalenii_PYR-1 RRGWKAVAAIDVP---------DVPDP--QALPSS-----APLLGIAYGY
Mb2192|M.bovis_AF2122/97 RRGWQAVAAVEV----TAAEQAEAAD--TTALPSAAELSNAPMLGVAYGY
Rv2170|M.tuberculosis_H37Rv RRGWQAVAAVEV----TAAEQAEAAD--TTALPSAAELSNAPMLGVAYGY
MAV_2324|M.avium_104 RRGWQGAAVVE----------AEVDY--DDQMPSAEELASAPLLGVAYGY
MLBr_00903|M.leprae_Br4923 RPGWQAVAAVEVRI--EVADVADMADGPAHPAPSADELNNAPLRGVAYGY
MMAR_3205|M.marinum_M RSGWKAVAAVDVPQTQPSTQLPDIIGADPNHELPAADLANAPMVGVAYGY
MUL_3513|M.ulcerans_Agy99 RSGWKAVAAVDVPQTQPSTQLPDIIGADPNHELPAADLANAPMVGVAYGY
MAB_1995c|M.abscessus_ATCC_199 RPGWQAAAIFDTP---------AAPSG--ITGPAEDLTEQARIVGIAYGY
* **:..* .: . * : *:****
MSMEG_4238|M.smegmatis_MC2_155 CGAPDQWWQQQVVSGLHRIGATREQIAELMSSYFELTELHIEPRAQGRGL
TH_0706|M.thermoresistible__bu CGAPDQWWQQQVVQGLHRVGVDRSRIAELTSSYFELTELHVAPRTQGRGL
Mflv_2977|M.gilvum_PYR-GCK CGAPDQWWQQQVISGLRRNGADADRISELMSSYFELTELHIHPRAQGHGL
Mvan_3534|M.vanbaalenii_PYR-1 CGSPDQWWQQQVVSGLKRSGVDGARITELMTSYFELTELHIHPQAQGRGL
Mb2192|M.bovis_AF2122/97 PGAPGQWWQQQVVLGLQRSGFPRLAIARLMTSYFELTELHILPRAQGRGL
Rv2170|M.tuberculosis_H37Rv PGAPGQWWQQQVVLGLQRSGFPRLAIARLMTSYFELTELHILPRAQGRGL
MAV_2324|M.avium_104 PGAPGQWWQQQVVLGLQRGGSPPQEIARLMNDYFELTELHIHPRAQGRGL
MLBr_00903|M.leprae_Br4923 PGAPGQWWQQQVVQGLQRSGLSTLEIARLMNSYFELTELHIHPHTQGRGI
MMAR_3205|M.marinum_M PGAPGQWWQQQVVLGLQRRGFPMHAIARVMSSYFELTELHIHPNAQGRGI
MUL_3513|M.ulcerans_Agy99 PGAPGQWWQQQVVLGLQRRGFPMHAIARVMSSYFELTELHIHPNAQGRGI
MAB_1995c|M.abscessus_ATCC_199 RGAADQWWHQQVSQGLRKTGLPRDRINALLNQYFELTELHVDPGLQGHGY
*:..***:*** **:: * * : ..********: * **:*
MSMEG_4238|M.smegmatis_MC2_155 GEALVRRLLADRGESHVLLSTPEINGEANRAWRLYRRLGFTDVIRGYHFA
TH_0706|M.thermoresistible__bu GEALARRLLADRPEAYVLLSTPEIEREANRAWRLYRRLGFTDVIRGYHFT
Mflv_2977|M.gilvum_PYR-GCK GEALIRRLLQDRTERQVLLSTPEINGEANRAWRLYRRLGFTDVIREYHFA
Mvan_3534|M.vanbaalenii_PYR-1 GEALIRRLLDGRSEQYVLLSTPEINGEGNRAWRLYRRLGFTDVIRGYHFA
Mb2192|M.bovis_AF2122/97 GEALARRLLAGRDEDNVLLSTPETNGEDNRAWRLYRRLGFTDIIRGYHFA
Rv2170|M.tuberculosis_H37Rv GEALARRLLAGRDEDNVLLSTPETNGEDNRAWRLYRRLGFTDIIRGYHFA
MAV_2324|M.avium_104 GEALARRLLAGRAEKNVLLSTPETNGEPNRAWRLYRRLGFTDIIRRYHFA
MLBr_00903|M.leprae_Br4923 GEALTRRLLAHRRENNVLLSTPETNGETNRAWRLYRRLGFMDIIRRHYFA
MMAR_3205|M.marinum_M GEALTRRLLAGRGEESVLLSTPESNGEDNRAWRLYRRLGFTDILRGYYFA
MUL_3513|M.ulcerans_Agy99 GEALTRRLLAGRGEESVLLSTPESNGEDNRAWRLYRRLGFTDILRGYYFA
MAB_1995c|M.abscessus_ATCC_199 GEALARRLLAGRTESHVLLSTPEISGESNRAWRLYRRLGFTDVIRQHQFA
**** **** * * ******* . * ************ *::* : *:
MSMEG_4238|M.smegmatis_MC2_155 GDPRAFAILGRPLPL------------------
TH_0706|M.thermoresistible__bu GDPRAFAILGRPLPL------------------
Mflv_2977|M.gilvum_PYR-GCK GDPRAFAILGRPLPLT-----------------
Mvan_3534|M.vanbaalenii_PYR-1 GDPRAFAILGRPLPL------------------
Mb2192|M.bovis_AF2122/97 GDPRAFAILGRTLPL------------------
Rv2170|M.tuberculosis_H37Rv GDPRAFAILGRTLPL------------------
MAV_2324|M.avium_104 GDPRPFAILGRELPL------------------
MLBr_00903|M.leprae_Br4923 GDPRAFAILGRTLPL------------------
MMAR_3205|M.marinum_M GDPRAFAILGRALPL------------------
MUL_3513|M.ulcerans_Agy99 GDPRAFAILGRALPL------------------
MAB_1995c|M.abscessus_ATCC_199 GDPRRFAVLGRALPLEPGYRQPGVSAVWHDNPR
**** **:*** ***