For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FLGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAVYPRDEFEKLARRASQASRSNPEARAFLRS LAAATDEQHPDAQGRITLSADHRRYANLSKDCVVIGSVDYLEIWDAQAWQEYQQAHEENFSAATDETLRD II*
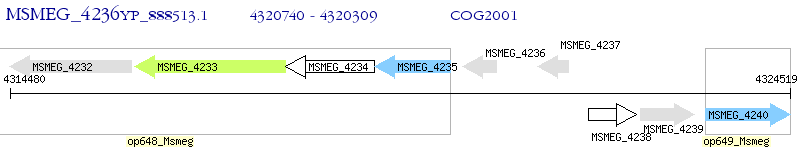
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4236 | mraZ | - | 100% (143) | cell division protein MraZ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2190c | - | 2e-70 | 88.73% (142) | cell division protein MraZ |
| M. gilvum PYR-GCK | Mflv_2979 | - | 1e-63 | 78.47% (144) | cell division protein MraZ |
| M. tuberculosis H37Rv | Rv2166c | - | 2e-70 | 88.73% (142) | cell division protein MraZ |
| M. leprae Br4923 | MLBr_00905 | - | 2e-71 | 88.81% (143) | cell division protein MraZ |
| M. abscessus ATCC 19977 | MAB_1997 | - | 9e-69 | 86.62% (142) | cell division protein MraZ |
| M. marinum M | MMAR_3203 | - | 4e-71 | 89.44% (142) | hypothetical protein MMAR_3203 |
| M. avium 104 | MAV_2327 | mraZ | 2e-72 | 90.21% (143) | cell division protein MraZ |
| M. thermoresistible (build 8) | TH_0704 | - | 1e-75 | 94.41% (143) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3511 | - | 3e-71 | 89.44% (142) | cell division protein MraZ |
| M. vanbaalenii PYR-1 | Mvan_3532 | - | 1e-62 | 77.08% (144) | cell division protein MraZ |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4236|M.smegmatis_MC2_155 --------MFLGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
TH_0704|M.thermoresistible__bu --------MFLGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
MLBr_00905|M.leprae_Br4923 --------MFLGTHTPKLDDKGRLTLPAKFRDALVGGLMVTKSQDHSLAV
MAV_2327|M.avium_104 --------MFLGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
Mb2190c|M.bovis_AF2122/97 --------MFLGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
Rv2166c|M.tuberculosis_H37Rv --------MFLGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
MMAR_3203|M.marinum_M MTLAGGGLVFLGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
MUL_3511|M.ulcerans_Agy99 --------MFLGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
MAB_1997|M.abscessus_ATCC_1997 --------MFLGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
Mflv_2979|M.gilvum_PYR-GCK --------MFFGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
Mvan_3532|M.vanbaalenii_PYR-1 --------MFFGTYTPKLDDKGRLTLPAKFRDALAGGLMVTKSQDHSLAV
:*:**:********************.***************
MSMEG_4236|M.smegmatis_MC2_155 YPRDEFEK-LARRASQASRSNPEARAFLRSLAAATDEQHPDAQGRITLSA
TH_0704|M.thermoresistible__bu YPRAEFEK-LARRASQASRSNPEARAFLRNLAAATDEQHPDSQGRITLSA
MLBr_00905|M.leprae_Br4923 YPRAEFEQ-LARRASKMSRSNPEARAFLRNLAAGTDEQHPDMQGRITLSA
MAV_2327|M.avium_104 YPRAEFEQ-LARRASKASKSNPDARAFLRNLAAGTDEQHPDAQGRITLSA
Mb2190c|M.bovis_AF2122/97 YPRAAFEQ-LARRASKAPRSNPEARAFLRNLAAGTDEQHPDSQGRITLSA
Rv2166c|M.tuberculosis_H37Rv YPRAAFEQ-LARRASKAPRSNPEARAFLRNLAAGTDEQHPDSQGRITLSA
MMAR_3203|M.marinum_M YPRSEFEQ-LARRASKAPRSNPEARAFLRNLAAGTDEQHPDSQGRITLSA
MUL_3511|M.ulcerans_Agy99 YPRSEFEQ-LARRASKAPRSNPEARAFLRNLAAGTDEQHPDSQGRITLSA
MAB_1997|M.abscessus_ATCC_1997 YPRAEFEQ-LARKAAAASRSNPEARAFLRNLAAATDEQHPDAQGRITLSA
Mflv_2979|M.gilvum_PYR-GCK HPRAEFEEMIAEISAKAKRGNPQARAYLRNLAASTDEQYPDAQGRITLSP
Mvan_3532|M.vanbaalenii_PYR-1 HPRAEFEEMIAEISAKAKRGNPQARAYLRNLAASTDEQYPDAQGRITLSA
:** **: :*. :: :.**:***:**.***.****:** *******.
MSMEG_4236|M.smegmatis_MC2_155 DHRRYANLSKDCVVIGSVDYLEIWDAQAWQEYQQAHEENFSAATDETLRD
TH_0704|M.thermoresistible__bu DHRRYANLTKDCVVIGAVDYLEIWDAQAWQEYQQMHEESFSAATDETLRD
MLBr_00905|M.leprae_Br4923 DHRRYANLSKDCVVIGAVDYLEIWDAQAWHDYQQTHEENFSAASDEALGD
MAV_2327|M.avium_104 DHRRYASLSKDCVVIGAVDYLEIWDAQAWQDYQQTHEENFSAASDEALGD
Mb2190c|M.bovis_AF2122/97 DHRRYASLSKDCVVIGAVDYLEIWDAQAWQNYQQIHEENFSAASDEALGD
Rv2166c|M.tuberculosis_H37Rv DHRRYASLSKDCVVIGAVDYLEIWDAQAWQNYQQIHEENFSAASDEALGD
MMAR_3203|M.marinum_M DHRRYAGLTKDCVVIGAVDYLEIWDAQAWHEYQQLHEENFSAASDEALGD
MUL_3511|M.ulcerans_Agy99 DHRRYAGLTKDCVVIGAVDYLEIWDAQAWHEYQQLHEENFSAASDEALGD
MAB_1997|M.abscessus_ATCC_1997 DHRRYADLSKDCVVIGSVDYLEIWDAAKWQQYLEEHEENFSTASDESLNG
Mflv_2979|M.gilvum_PYR-GCK EHRRYANLTKDCVVTGSIDFLEIWDAQAWQEYQELHEENFSAASDEALGD
Mvan_3532|M.vanbaalenii_PYR-1 EHRRYANLTKECVVTGSIGFLEIWDAQAWQDYQELHEENFSAASDEALGD
:*****.*:*:*** *::.:****** *::* : ***.**:*:**:* .
MSMEG_4236|M.smegmatis_MC2_155 II
TH_0704|M.thermoresistible__bu II
MLBr_00905|M.leprae_Br4923 II
MAV_2327|M.avium_104 II
Mb2190c|M.bovis_AF2122/97 IF
Rv2166c|M.tuberculosis_H37Rv IF
MMAR_3203|M.marinum_M IF
MUL_3511|M.ulcerans_Agy99 IF
MAB_1997|M.abscessus_ATCC_1997 IF
Mflv_2979|M.gilvum_PYR-GCK IL
Mvan_3532|M.vanbaalenii_PYR-1 IL
*: