For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAQIAPLVFDAEGHPDPMTLTDLEQRLPGLTDLVIFSHGWNNDEAAATLLYDRWFRLLAPHLDPARNVGF VGVRWPSTLWRDEPIPDFPATPAGGAEGAAALGGAREVAADSDAGNPALDPQELAELKQLFPTAEQQLDT LAALLAQPPDVDAVPDLFEALQDFHKATPDGFDDGETEDTQDGPGMVADDQDPAELFNTFADQLLSAGVQ FGDDGTGAAGLGDFAGKLWRGAKEALRQLSYWKMKNRAGVVGKNGLGPVIDKLARSAPGLRIHLVGHSFG ARLVCYALAGMSGAQPSPVKSVTLLQGAFSRFSFIDRLPFRDGAGALAGLLGRVDGPLTVCFSRHDRALS TFYPLASAAVGDDAAGLDDPLFRWRAMGSHGAFRSESLGLGAVGAQYPFEAGKILNLDASEVVKADAGPS GAHSDIFHDELAWVVATAGGLN*
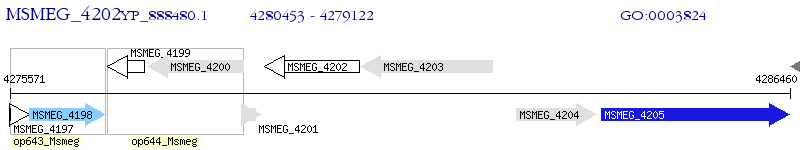
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4202 | - | - | 100% (443) | hypothetical protein MSMEG_4202 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3911 | - | 1e-66 | 39.47% (418) | hypothetical protein MAB_3911 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4202|M.smegmatis_MC2_155 MTAQIAPLVFDAEGHPDPMTLTD------LEQRLP--GLTDLVIFSHGWN
MAB_3911|M.abscessus_ATCC_1997 MEPINGLRRFEVQFNADGSFNTEDGGDGGLAAAVATGGITDLYVMAHGWN
* . . *:.: :.* *: * :. *:*** :::****
MSMEG_4202|M.smegmatis_MC2_155 NDEAAATLLYDRWFRLLAPHLDPARN-VGFVGVRWPSTLWRDEPIPDFPA
MAB_3911|M.abscessus_ATCC_1997 NGMASARALYDAMFGLLAGQLGDRRATSAAVGIIWPSILFPD----DDPA
*. *:* *** * *** :*. * . **: *** *: * * **
MSMEG_4202|M.smegmatis_MC2_155 TPAGGAEGAAALGGAREVAADSDAGNPALDPQELAELKQLFPTAEQQLDT
MAB_3911|M.abscessus_ATCC_1997 TAS---------------------AQPASPTQVTAALAATMPDQKRTLDE
*.: .:** .* * * :* :: **
MSMEG_4202|M.smegmatis_MC2_155 LAALLAQPPDVDAVPDLFEALQDFHKATPDGFDDGETEDTQDGPGMVADD
MAB_3911|M.abscessus_ATCC_1997 LGRQLEIQPQDPVELKKFVTLATGLVTTAP-----QSEE----------D
*. * *: . . * :* :*. ::*: *
MSMEG_4202|M.smegmatis_MC2_155 QDPAELFNTFADQLLSAGVQFGDDGTG-AAGLGDFAGKLWRGAKEALRQL
MAB_3911|M.abscessus_ATCC_1997 SGEAAILDSDAMKVLGHAATLAPRPTGNAQGSGNPFTRLWSGARELLRTM
.. * :::: * ::*. .. :. ** * * *: :** **:* ** :
MSMEG_4202|M.smegmatis_MC2_155 SYWKMKNRAGVVGKNGLGPVIDKLARSAPGLRIHLVGHSFGARLVCYALA
MAB_3911|M.abscessus_ATCC_1997 SYYEMKNRAGVVGERGLGPLLGSLSGPDGPPRIHLIGHSFGARLVSYALA
**::*********:.****::..*: . ****:*********.****
MSMEG_4202|M.smegmatis_MC2_155 GMSGAQPSPVKSVTLLQGAFSRFSFIDRLPFR-DGAGALAGLLGRVDGPL
MAB_3911|M.abscessus_ATCC_1997 GLPATGTSPVKSLTLIQGAFSHFAFAHSLPFDIDRHGGLSGLEARVDGPL
*:..: .*****:**:*****:*:* . *** * *.*:** .******
MSMEG_4202|M.smegmatis_MC2_155 TVCFSRHDRALSTFYPLASAAVGDDAAGLDDPLFRWRAMGSHGAFR---S
MAB_3911|M.abscessus_ATCC_1997 LATFTAADRAVGWWYPAASALARQDSQAAQELTFRWGAMGHDGYQQDPAA
. *: ***:. :** *** . :*: . :: *** *** .* : :
MSMEG_4202|M.smegmatis_MC2_155 ESLGLGAVGAQYPFEAGKILNLDASEVVKADAGP-SGAHSDIFHDELAWV
MAB_3911|M.abscessus_ATCC_1997 TTVMLKEAGKPYAFERGRFYALDANAVIAADESPFSGAHSDIKHPEVTWA
:: * .* *.** *:: ***. *: ** .* ******* * *::*.
MSMEG_4202|M.smegmatis_MC2_155 VATAGGLN
MAB_3911|M.abscessus_ATCC_1997 IAAAAQAA
:*:*.