For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLDVLGHLVEVDRNRCEGHAQCVATAPGFFTIDENGELGATGTDTATSAVELEHVEMAVSACPMNALRLI EPRPADPSHAPE*
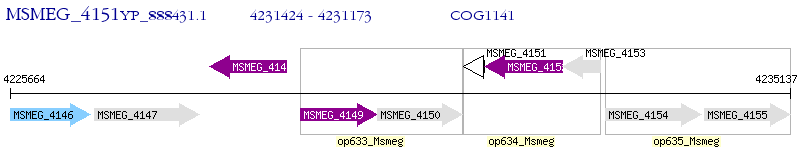
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4151 | - | - | 100% (83) | hypothetical protein MSMEG_4151 |
| M. smegmatis MC2 155 | MSMEG_5864 | - | 8e-07 | 34.92% (63) | cytochrome P450 51 |
| M. smegmatis MC2 155 | MSMEG_6694 | - | 1e-06 | 39.34% (61) | hypothetical protein MSMEG_6694 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0786c | - | 5e-07 | 34.33% (67) | ferredoxin |
| M. gilvum PYR-GCK | Mflv_1595 | - | 3e-06 | 36.51% (63) | hypothetical protein Mflv_1595 |
| M. tuberculosis H37Rv | Rv0763c | - | 5e-07 | 34.33% (67) | ferredoxin |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1213c | - | 2e-06 | 31.34% (67) | putative ferredoxin |
| M. marinum M | MMAR_4730 | - | 1e-05 | 32.43% (74) | ferredoxin |
| M. avium 104 | MAV_0924 | - | 3e-07 | 35.62% (73) | hypothetical protein MAV_0924 |
| M. thermoresistible (build 8) | TH_0529 | - | 7e-07 | 32.91% (79) | conserved domain protein |
| M. ulcerans Agy99 | MUL_0472 | - | 1e-05 | 29.23% (65) | ferredoxin |
| M. vanbaalenii PYR-1 | Mvan_5162 | - | 2e-07 | 37.88% (66) | hypothetical protein Mvan_5162 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1595|M.gilvum_PYR-GCK -----MG---CYRVELDEDLCQGHAMCELEAPDVFRVPKRGVVEILDSEP
Mvan_5162|M.vanbaalenii_PYR-1 -----MSGNRGYRVHVDEDLCQGHAMCELEAPDVFRVPKRGVVEITDPEP
MAB_1213c|M.abscessus_ATCC_199 -----MA----YRVDVDTDLCQGHAMCELEAPDYFRVPKRGTVEILNNEP
Mb0786c|M.bovis_AF2122/97 -----MG----YRVEADRDLCQGHAMCELEAPEYFRVPKRGQVEILDPEP
Rv0763c|M.tuberculosis_H37Rv -----MG----YRVEADRDLCQGHAMCELEAPEYFRVPKRGQVEILDPEP
MUL_0472|M.ulcerans_Agy99 -----MS----YRIEADLDLCQGHAMCELEAPDYFRVPKRGKVEIIDPEP
MSMEG_4151|M.smegmatis_MC2_155 MTLDVLG----HLVEVDRNRCEGHAQCVATAPGFFTIDENGELGATGTDT
MMAR_4730|M.marinum_M -----------MRIRLDRTVCDGFGLCAKQAPGYFTLDDWGYACLSGDGT
MAV_0924|M.avium_104 -----------MRIRLDRTVCDGFGICAKHAPGYFSLDDWGYASLIGDGT
TH_0529|M.thermoresistible__bu -----------MRIRLDRTICDGFGVCAKHAPGYFSLDDWGYASLIGDGT
: * *:*.. * ** * : . * . .
Mflv_1595|M.gilvum_PYR-GCK PDELR--EAVEMAVEMCPTRALTITEKD----------------------
Mvan_5162|M.vanbaalenii_PYR-1 PDELR--EAVELAVEMCPTRALTIIEKE----------------------
MAB_1213c|M.abscessus_ATCC_199 PDDAR--DEIERAVEECPARALFITEK-----------------------
Mb0786c|M.bovis_AF2122/97 PEEAR--GVIKHAVWACPTQALSIRETGE---------------------
Rv0763c|M.tuberculosis_H37Rv PEEAR--GVIKHAVWACPTQALSIRETGE---------------------
MUL_0472|M.ulcerans_Agy99 PEQAR--PEIEQAVRMCPTQALSIKAKED---------------------
MSMEG_4151|M.smegmatis_MC2_155 ATSAVELEHVEMAVSACPMNALRLIEPRPADPSHAPE-------------
MMAR_4730|M.marinum_M VREEDR-DAVLRALMDCPAHAITEIGERQPDVVQHSPASAEDPAEHLKTE
MAV_0924|M.avium_104 VAEADR-DAVMRALLDCPVHAIAEIGERTAPSHRPPLAEATDPAEHLKTE
TH_0529|M.thermoresistible__bu VPEQDR-DAVMRALLDCPVHAIIKIADNRPEEPPASPASAPE--------
. : *: ** .*:
Mflv_1595|M.gilvum_PYR-GCK ----------
Mvan_5162|M.vanbaalenii_PYR-1 ----------
MAB_1213c|M.abscessus_ATCC_199 ----------
Mb0786c|M.bovis_AF2122/97 ----------
Rv0763c|M.tuberculosis_H37Rv ----------
MUL_0472|M.ulcerans_Agy99 ----------
MSMEG_4151|M.smegmatis_MC2_155 ----------
MMAR_4730|M.marinum_M SNEAQWGFTR
MAV_0924|M.avium_104 ENEAEWGFTR
TH_0529|M.thermoresistible__bu -NETVGGLR-