For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTADTVRVPPNPVSDNAFLADFAAMSTFGATPAGGVERQAGTTADGENRRWLAAWLAERGFEVRYDRIGN MFGLYELVPGAPYVLAGSHLDSQPRGGRYDGAYGVLAAAHAATRLLDHFRANPGAARYNIAVVNWFNEEG SRFKPSMQGSGVFTGKLALQEALDTTDVDGVSVAEALGALDMLGERDVFTVGDHHTAGGVQVASYAEIHI EQGRELDKAGVPIGIVPSTWGANKYEISVVGAQSHTGATAIADRQDALLGAAKVVVALRELADSYGEDLH TSCGQLTVYPNSPVVVAREVHMHLDLRSPSDQLLDEADARLNRTLAEIEVAADVHIERRFAHTWKGHTYT PEGVTLAEAAIEALELNSMHVRTRAGHDSTNMKDVVPTVMLFIPSVDGVSHAENEFTHDKDLTTGVDVLT ETMARMVEGDL
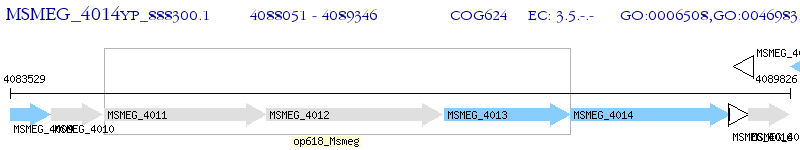
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4014 | - | - | 100% (431) | N-carbamoyl-L-amino acid amidohydrolase |
| M. smegmatis MC2 155 | MSMEG_3995 | - | e-155 | 63.34% (431) | N-carbamoyl-L-amino acid amidohydrolase |
| M. smegmatis MC2 155 | MSMEG_0819 | - | e-106 | 48.82% (424) | N-carbamoyl-L-amino acid amidohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |