For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIGCHYRRLGTDGEYQLFESTPDTRSNWDESIQHGSPPLALLTKAIEELMAGSGLRVGRLTLDILGAIPV APVRVRTWVERPGSRISLAVAEMAAARPDGEWRAVAKVSAWLLATGDTSDVTTDRHPPLVEGEPADVPHN WVGAPGYLDTVTWRRQATPEGASAEVWMSPLTPVVDDEETTALQRLALVVDSANGVGAALDPKKFIFMNT DTTVHLHRLPEGSDFAVRARGSIGPDGIGVTTAEIFDRKGFIGTSAQTLLVMRAP
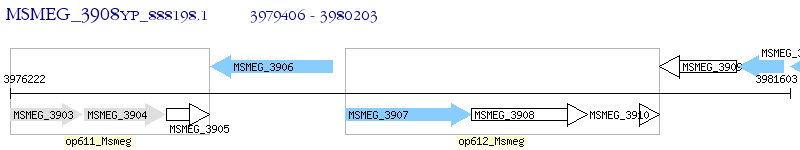
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3908 | - | - | 100% (265) | hypothetical protein MSMEG_3908 |
| M. smegmatis MC2 155 | MSMEG_2560 | - | 1e-47 | 40.71% (253) | hypothetical protein MSMEG_2560 |
| M. smegmatis MC2 155 | MSMEG_3458 | - | 8e-16 | 32.46% (228) | hypothetical protein MSMEG_3458 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3063 | - | 1e-114 | 75.29% (263) | hypothetical protein Mflv_3063 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2154c | - | 2e-81 | 57.14% (266) | hypothetical protein MAB_2154c |
| M. marinum M | MMAR_3096 | - | 1e-95 | 64.39% (264) | hypothetical protein MMAR_3096 |
| M. avium 104 | MAV_5133 | - | 2e-45 | 38.32% (274) | hypothetical protein MAV_5133 |
| M. thermoresistible (build 8) | TH_0042 | - | 1e-103 | 68.70% (262) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2342 | - | 1e-94 | 64.02% (264) | hypothetical protein MUL_2342 |
| M. vanbaalenii PYR-1 | Mvan_3464 | - | 1e-110 | 74.42% (258) | hypothetical protein Mvan_3464 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3063|M.gilvum_PYR-GCK ----MIECLYRRIG-ADGEYTVFESTDGTRSNWDPQIQHGSPPLALLTRA
Mvan_3464|M.vanbaalenii_PYR-1 ----MIESLFRRLP-ADGEYQVFESTDGTRSNWDPQIQHGSPPLALLTRA
MSMEG_3908|M.smegmatis_MC2_155 ----MIGCHYRRLG-TDGEYQLFESTPDTRSNWDESIQHGSPPLALLTKA
TH_0042|M.thermoresistible__bu ----VTRFYYRRLR-TDGEFEVLESTGATGSNWDAGIQHGSPPLALLTKL
MMAR_3096|M.marinum_M ----MNGCYYRRLPGADGDYQMFDSTDFTRSNWDPAIQHGSPPLALLTKL
MUL_2342|M.ulcerans_Agy99 ----MNGCYYRRLPGADGDYQMFDSTDFTRSNWDPDIQHGSPPLALLTKL
MAB_2154c|M.abscessus_ATCC_199 MTLDLTGCYYRRLD-ADGEFQVFESTELTASGWGPNLQHGSPPMALLLKA
MAV_5133|M.avium_104 ----MTDAYYELID-SDASGERFRATDLARGTWSATIQHGGPVSALLTRA
: :. : :*.. : :* : . *. :***.* *** :
Mflv_3063|M.gilvum_PYR-GCK IEDLAEGTG---LRVGRLTLDILGAIPVT-TVHVRSWVERPGSRISLMAA
Mvan_3464|M.vanbaalenii_PYR-1 IEELAAGTG---LRVGRLTLDILGAIPVS-TVRVRARVDRPGARISLMTA
MSMEG_3908|M.smegmatis_MC2_155 IEELMAGSG---LRVGRLTLDILGAIPVA-PVRVRTWVERPGSRISLAVA
TH_0042|M.thermoresistible__bu VEDSLDGTTGPRLRVGRLVMDILGPIPVA-PVRVRAWTERPGSRISLRTA
MMAR_3096|M.marinum_M IEELSAGSG---LRIGRLSLDILGAIPVA-PVRARAWVQRPGSRVAMMVA
MUL_2342|M.ulcerans_Agy99 IEELSAGSG---LRIGRFSLDILGAIPVA-PVRARAWVQRPGSRVAMMVA
MAB_2154c|M.abscessus_ATCC_199 IEELPEPREN--LRIGRVALDILGPVPLE-PVRVRAWIERPGRRIALAVA
MAV_5133|M.avium_104 LERCEQRDD---TRLSRVVIDLLGGVPVEGDLWVRAEVQRPGKQIELLGA
:* *:.*. :*:** :*: : .*: :*** :: : *
Mflv_3063|M.gilvum_PYR-GCK EMLVDRPDGPRRAVARVTAWLLAPSDTADAVTDRHP-PLIEGEASTVAHA
Mvan_3464|M.vanbaalenii_PYR-1 EMLTDRPDGTRRAVARVTAWLLAPSDTGDAATDRYP-PLVEGEAMTVAHP
MSMEG_3908|M.smegmatis_MC2_155 EMAAARPDGEWRAVAKVSAWLLATGDTSDVTTDRHP-PLVEGEPADVPHN
TH_0042|M.thermoresistible__bu EMQAQRPDGSWRAVARLSAWLLAVSDTADVATDRHP-PLQEGPVVGEPHA
MMAR_3096|M.marinum_M EMVADR------VLARVTAWLLAVSDTTDVASDRYP-PLVEGPAQPRPAA
MUL_2342|M.ulcerans_Agy99 EMVADR------VLARVTAWLLAVSDTTDVASDRYP-PLVEGPAQPRPAA
MAB_2154c|M.abscessus_ATCC_199 EMEARANGG-YRAVARASAWLLATSVTADKASDRFP-PLPEYPAGPTPQA
MAV_5133|M.avium_104 QMLAPGPDGAPRPVARASGWRLQHQDTGSLAHAAAPLPRPRAEAYDRNLK
:* . :*: :.* * * . . * * .
Mflv_3063|M.gilvum_PYR-GCK WEGAPG-YLETVSWRRQQD-QDG-AAVAWLSPLVPLVDSEPTTALQRLAM
Mvan_3464|M.vanbaalenii_PYR-1 WEGAPG-YLETVSWRRQFT-ADGEAAVSWLSPLVPLVDAEPTTPLQRLAM
MSMEG_3908|M.smegmatis_MC2_155 WVGAPG-YLDTVTWRRQAT-PEGASAEVWMSPLTPVVDDEETTALQRLAL
TH_0042|M.thermoresistible__bu WHGAPG-YLETIDFRRQES-AGESGTVAWMTPLARVVDTDDPTPLQRLAM
MMAR_3096|M.marinum_M FAGVGG-YFDALSWRPQDT-EGQPAAVSWFRPTAHIVDTDPASSLQQLAA
MUL_2342|M.ulcerans_Agy99 FAGVGG-YFDALSWRPQDT-EGQPAAVSWFRPTAHIVDTDPASPLQQLAA
MAB_2154c|M.abscessus_ATCC_199 FGFEGSNYSKSVDFQWFTV-PNGEPQVAWMRPHVHIVDTEPTTPWQRLAS
MAV_5133|M.avium_104 ARDWDRNYVHSLQWLWLTEPLSEGPGESWISPTVDLVKGESMTRLQRLFA
* .:: : *: * . :*. : : *:*
Mflv_3063|M.gilvum_PYR-GCK VVDSANGIGAALDPQRFLFMNTDTAVHLHRLPEGQDFAVRARGSIGPDGI
Mvan_3464|M.vanbaalenii_PYR-1 VVDSANGIGAAVDPRRFVFMNTDTVVHLHRLPEGNDFGLRARGSIGPDGV
MSMEG_3908|M.smegmatis_MC2_155 VVDSANGVGAALDPKKFIFMNTDTTVHLHRLPEGSDFAVRARGSIGPDGI
TH_0042|M.thermoresistible__bu VVDSANGIGAVLDPNRFVFMNTDTTVHLHRLPVGDDFAVRARASIGPDGI
MMAR_3096|M.marinum_M VVDCANGVGAILDPNRFLFMNTDTVVHLHRLPTGTDFALRARASVGPDGL
MUL_2342|M.ulcerans_Agy99 VVDCANGVGAILDPNRFLFMNTDTVVHLHRLPTGTDFALRARASVGPDGL
MAB_2154c|M.abscessus_ATCC_199 VIDSANGIGAVLNPLRFVYMNTDTVMHVHRIPQGDEFGVRARMSVGPDGV
MAV_5133|M.avium_104 VADCANGIGSKLDITKWTFVNTDLAVHVFRVPDGDWIGIRAETSYGPDGI
* *.***:*: :: :: ::*** .:*:.*:* * :.:**. * ****:
Mflv_3063|M.gilvum_PYR-GCK GVTTAELFDRNGFVGTSAQTLLVQRLG---
Mvan_3464|M.vanbaalenii_PYR-1 GVTTAEIFDRAGFIGTSAQTLLVQRRG---
MSMEG_3908|M.smegmatis_MC2_155 GVTTAEIFDRKGFIGTSAQTLLVMRAP---
TH_0042|M.thermoresistible__bu GVTTAEVFDRQGFVGTCAQTLLVQRLG---
MMAR_3096|M.marinum_M GVTTAEIFDKTGFVGTSAQTLLVRRR----
MUL_2342|M.ulcerans_Agy99 GVTTAEIFDTTGFVGTSAQTLLVRRR----
MAB_2154c|M.abscessus_ATCC_199 GVTTAEIFDRQGHIGSSAQTVLVQRR----
MAV_5133|M.avium_104 GTTIGTLFDEQGAVGAIQQSVLVRRRPGRA
*.* . :** * :*: *::** *