For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGNRPTGPFAEGDRVQLTDAKGRHYTMVLNHGGEFHTHRGIIAFDDVIGLPEGSVVKSTNGDQFLVLRPL LVDYVMSMPRGAQVIYPKDAAQIVHEGDIFPGARVLEAGAGSGALTCSLLRAVGPEGSVTSYEVRDDHAV HAERNVVTFFGERPANWNLVIGDVSDYDGPEMDRVVLDMLAPWEVLPAVSRALVAGGVLMIYVATVTQLS RAVEALREQQCWTEPRAWESMQRGWHVVGLAVRPQHTMRGHTAFLISARKLAPGAVAPMPLRRKRQIGGG I*
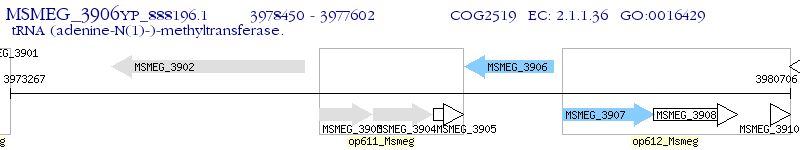
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3906 | - | - | 100% (282) | tRNA (adenine-N(1)-)-methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2142c | - | 1e-119 | 77.29% (273) | RNA methyltransferase |
| M. gilvum PYR-GCK | Mflv_3065 | - | 1e-142 | 85.87% (283) | tRNA (adenine-N(1)-)-methyltransferase |
| M. tuberculosis H37Rv | Rv2118c | - | 1e-119 | 77.29% (273) | RNA methyltransferase |
| M. leprae Br4923 | MLBr_01313 | - | 1e-118 | 75.18% (278) | hypothetical protein MLBr_01313 |
| M. abscessus ATCC 19977 | MAB_2159 | - | 1e-130 | 80.88% (272) | hypothetical protein MAB_2159 |
| M. marinum M | MMAR_3094 | - | 1e-121 | 77.29% (273) | RNA methyltransferase |
| M. avium 104 | MAV_2397 | - | 1e-120 | 77.01% (274) | PimT protein |
| M. thermoresistible (build 8) | TH_0044 | - | 1e-140 | 87.13% (272) | POSSIBLE RNA METHYLTRANSFERASE |
| M. ulcerans Agy99 | MUL_2340 | - | 1e-119 | 76.87% (268) | RNA methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_3462 | - | 1e-140 | 87.13% (272) | tRNA (adenine-N(1)-)-methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3094|M.marinum_M -----------MSATGPFHVGERVQLTDAKGRHYTITLTPGAEFHTHRGR
MUL_2340|M.ulcerans_Agy99 -----------MSATGPFHVGERVQLTDAKGRHYTITLTPGAEFHTHRGR
Mb2142c|M.bovis_AF2122/97 -----------MSATGPFSIGERVQLTDAKGRRYTMSLTPGAEFHTHRGS
Rv2118c|M.tuberculosis_H37Rv -----------MSATGPFSIGERVQLTDAKGRRYTMSLTPGAEFHTHRGS
MLBr_01313|M.leprae_Br4923 -----------MSATGPFTVGERVQLTDTKGRHYTMSLTPGGEFHTHRGS
MAV_2397|M.avium_104 -----------MSVTGPFIVGERVQLTDAKGRHYTMVLAAGAEFHTHRGS
Mflv_3065|M.gilvum_PYR-GCK MAARLVAGSSAVRRTGPFAVGDRVQLTDAKGRRYTMLLKPGGEFHTHRGI
Mvan_3462|M.vanbaalenii_PYR-1 -----------MRRTGPFDVGDRVQLTDAKGRRYTMLLKPGGEFHTHRGI
MSMEG_3906|M.smegmatis_MC2_155 -----MAGN---RPTGPFAEGDRVQLTDAKGRHYTMVLNHGGEFHTHRGI
TH_0044|M.thermoresistible__bu -----------VSRTGPFAVGDRVQLTDAKGRRYTMVLKPGGEFHTHRGV
MAB_2159|M.abscessus_ATCC_1997 -----------MSRTGPFIVGDRVQLTDAKGRHYTMVLEPGKEFHTHRGA
**** *:******:***:**: * * *******
MMAR_3094|M.marinum_M IAHDVIIGLEQGSVVKSSNGALFLVLRPLLVDYVMSMPRGPQVIYPKDAA
MUL_2340|M.ulcerans_Agy99 IAHDVIIGLEQGSVVESSNGALFLVLRPLLVDYVMSMPRGPQVIYPKDAA
Mb2142c|M.bovis_AF2122/97 IAHDAVIGLEQGSVVKSSNGALFLVLRPLLVDYVMSMPRGPQVIYPKDAA
Rv2118c|M.tuberculosis_H37Rv IAHDAVIGLEQGSVVKSSNGALFLVLRPLLVDYVMSMPRGPQVIYPKDAA
MLBr_01313|M.leprae_Br4923 IAHDGIIGLEQGSVVKSSTGALFLVLRPLLIDYVMSMPRGPQVIYPKDAA
MAV_2397|M.avium_104 IPHDAVIGLEQGSVVKSSNGAVYLVMRPLLIDYVMSMPRGPQVIYPKDAA
Mflv_3065|M.gilvum_PYR-GCK IALDDVVGLPEGSVIKSTNGDQFLVLRPLLVDYVMSMPRGAQVVYPKDAA
Mvan_3462|M.vanbaalenii_PYR-1 IALDEVIGLPEGSVVKSTNGDQFLVLRPLLVDYVMSMPRGAQVVYPKDAA
MSMEG_3906|M.smegmatis_MC2_155 IAFDDVIGLPEGSVVKSTNGDQFLVLRPLLVDYVMSMPRGAQVIYPKDAA
TH_0044|M.thermoresistible__bu VALDDVIGLPEGSVVTASNGDKFLVMRPLLTDFVLSMPRGPQVIYPKDAA
MAB_2159|M.abscessus_ATCC_1997 IVHDEVIGIPEGSVVKSTNGDQFLVLRPLLLDYVLSMPRGAQVIYPKDAA
: * ::*: :***: ::.* :**:**** *:*:*****.**:******
MMAR_3094|M.marinum_M QIVHEGDIFPGARVLEAGAGSGALTLSLLRAVGPQGQVISYEQRADHAEH
MUL_2340|M.ulcerans_Agy99 QIVHEGDIFPGARVLDAGAGSGALTLSLLRAVGPQGQVISYEQRADHAEH
Mb2142c|M.bovis_AF2122/97 QIVHEGDIFPGARVLEAGAGSGALTLSLLRAVGPAGQVISYEQRADHAEH
Rv2118c|M.tuberculosis_H37Rv QIVHEGDIFPGARVLEAGAGSGALTLSLLRAVGPAGQVISYEQRADHAEH
MLBr_01313|M.leprae_Br4923 QIVHEGDIFPGARVLEAGAGSGALTCSLLRAVGPQGQVISYELRTDHAEH
MAV_2397|M.avium_104 QIVHEGDIFPGARVLEAGAGSGALTCSLLRAVGPQGRVISYEQRADHAEH
Mflv_3065|M.gilvum_PYR-GCK QIVHEGDIFPGARVLEAGAGSGALTCSLLRAVGPDGTVTSYEVRDDHAEH
Mvan_3462|M.vanbaalenii_PYR-1 QIVHEGDIFPGARVLEAGAGSGALTCSLLRAVGPQGRVTSYEVREDHAEH
MSMEG_3906|M.smegmatis_MC2_155 QIVHEGDIFPGARVLEAGAGSGALTCSLLRAVGPEGSVTSYEVRDDHAVH
TH_0044|M.thermoresistible__bu QIVHEGDIFPGARVLEAGAGSGALTCSLLRAVGPEGSVTSYEVRDDHAEH
MAB_2159|M.abscessus_ATCC_1997 QIVHDGDIFPGARVLEAGAGSGALTCSLLRAVGPQGAVISYEIREDHAEH
****:**********:********* ******** * * *** * *** *
MMAR_3094|M.marinum_M ARRNVTNFY------GEAPDNWQLIISDIADSELPDGSIDRVVLDMLAPW
MUL_2340|M.ulcerans_Agy99 ARRNVTNFY------GEAPENWQLIISDIADSELPDGSIDRVVLDMLAPW
Mb2142c|M.bovis_AF2122/97 ARRNVSGCY------GQPPDNWRLVVSDLADSELPDGSVDRAVLDMLAPW
Rv2118c|M.tuberculosis_H37Rv ARRNVSGCY------GQPPDNWRLVVSDLADSELPDGSVDRAVLDMLAPW
MLBr_01313|M.leprae_Br4923 ARRNVSNFFCVSGAEGRLSDNWQLIVSDVANSELPDGSVDRVVLDMLAPW
MAV_2397|M.avium_104 AQRNVSVFFG-----GEAPDNWQLIVDDVADSDLPDRSVDRAVLDMLAPW
Mflv_3065|M.gilvum_PYR-GCK AIRNVETFF------GERPANWELVIADLSG--YDGPEVDRVVLDMLSPW
Mvan_3462|M.vanbaalenii_PYR-1 AQRNVATFF------GEQPPNWDLVIADVAD--YDGPEVDRVVLDMLAPW
MSMEG_3906|M.smegmatis_MC2_155 AERNVVTFF------GERPANWNLVIGDVSD--YDGPEMDRVVLDMLAPW
TH_0044|M.thermoresistible__bu AVRNVTTFF------GERPANWELVVADLAD--YAGGEVDRVVLDMLAPW
MAB_2159|M.abscessus_ATCC_1997 AVRNVTTFF------GERPDNWDLVIGDLAERPADAGSVDRVVLDMLAPW
* *** : *. . ** *:: *:: .:**.*****:**
MMAR_3094|M.marinum_M EVLDSVSRLVVAGGVLMIYVATVTQLSKVVEAVRTQQCWTEPRSWETLQR
MUL_2340|M.ulcerans_Agy99 EVLDPVSRLVVAGGVLMIYVATVTQLSKVVEAVRTQQCWTEPRSWETLQR
Mb2142c|M.bovis_AF2122/97 EVLDAVSRLLVAGGVLMVYVATVTQLSRIVEALRAKQCWTEPRAWETLQR
Rv2118c|M.tuberculosis_H37Rv EVLDAVSRLLVAGGVLMVYVATVTQLSRIVEALRAKQCWTEPRAWETLQR
MLBr_01313|M.leprae_Br4923 EVLDTVSRLVIAGGVLVIYVATVTQLSRAVEALRVQQCWVEPRAWETLQR
MAV_2397|M.avium_104 EVLESVARLLIPGGVLMIYVATVTQLSRAVEALRAQQCWTEPRSWETLQR
Mflv_3065|M.gilvum_PYR-GCK EVLPAVAEALVPGGVVMIYVATVTQLSKTVEAMREQQCWTEPRAWESMQR
Mvan_3462|M.vanbaalenii_PYR-1 DVLGAVGKALVPGGVLMIYVATVTQLSKTVEALREQQCWTEPRAWESMQR
MSMEG_3906|M.smegmatis_MC2_155 EVLPAVSRALVAGGVLMIYVATVTQLSRAVEALREQQCWTEPRAWESMQR
TH_0044|M.thermoresistible__bu EVLDAVARALVAGGVLMVYVATVTQLSRIVEALREQQCWTEPRAWETMQR
MAB_2159|M.abscessus_ATCC_1997 ETLPAVAETLVPGGVLIVYVATVTQLSRVVEALREQQCWTEPRSWETMQR
:.* .*.. ::.***:::*********: ***:* :***.***:**::**
MMAR_3094|M.marinum_M GWNVVGLAVRPQHSMRGHTAFLVAARRLAPGAVAPAPLG-----------
MUL_2340|M.ulcerans_Agy99 GWNVVGLAVRPQHSMRGHTAFLVAARRLAPGAVAPAPLGPQTRGARWLAG
Mb2142c|M.bovis_AF2122/97 GWNVVGLAVRPQHSMRGHTAFLVATRRLAPGAVAPAPLG-----------
Rv2118c|M.tuberculosis_H37Rv GWNVVGLAVRPQHSMRGHTAFLVATRRLAPGAVAPAPLG-----------
MLBr_01313|M.leprae_Br4923 GWNVVGLAVRPEHLMRGHTAFLILARRLAPGAVAPAPWG-----------
MAV_2397|M.avium_104 GWNVVGLAVRPQHSMRGHTAFLIFTRRLAPGVIAPAPLG-----------
Mflv_3065|M.gilvum_PYR-GCK GWHVVGLAVRPEHTMRGHTAFLISARRLAPGTVTPVPLR-----------
Mvan_3462|M.vanbaalenii_PYR-1 GWHVVGLAVRPEHSMRGHTAFLISARRLAPGTVAPIPLR-----------
MSMEG_3906|M.smegmatis_MC2_155 GWHVVGLAVRPQHTMRGHTAFLISARKLAPGAVAPMPLR-----------
TH_0044|M.thermoresistible__bu GWHVVGLAVRPEHTMRGHTAFLVSARKLAPGAVAPTPLR-----------
MAB_2159|M.abscessus_ATCC_1997 GWNVVGLAVRPEHSMRGHTAFLVSARRLAPGTITPTPLR-----------
**:********:* ********: :*:****.::* *
MMAR_3094|M.marinum_M --------------------------------RKREGRDG----------
MUL_2340|M.ulcerans_Agy99 NRPNRASPRCGAGRGAPTTVRTRGARLPGVRPRRGHQRHGCARWKPRPPQ
Mb2142c|M.bovis_AF2122/97 --------------------------------RKREGRDG----------
Rv2118c|M.tuberculosis_H37Rv --------------------------------RKREGRDG----------
MLBr_01313|M.leprae_Br4923 --------------------------------RKR---------------
MAV_2397|M.avium_104 --------------------------------RKREGRDG----------
Mflv_3065|M.gilvum_PYR-GCK --------------------------------KKRVTGG-----------
Mvan_3462|M.vanbaalenii_PYR-1 --------------------------------KKRLS-------------
MSMEG_3906|M.smegmatis_MC2_155 --------------------------------RKRQIGGGI---------
TH_0044|M.thermoresistible__bu --------------------------------RKRQI-------------
MAB_2159|M.abscessus_ATCC_1997 --------------------------------RKRLPS------------
::
MMAR_3094|M.marinum_M ---------------------
MUL_2340|M.ulcerans_Agy99 FRPVGAARVIRSRRPTRRTCA
Mb2142c|M.bovis_AF2122/97 ---------------------
Rv2118c|M.tuberculosis_H37Rv ---------------------
MLBr_01313|M.leprae_Br4923 ---------------------
MAV_2397|M.avium_104 ---------------------
Mflv_3065|M.gilvum_PYR-GCK ---------------------
Mvan_3462|M.vanbaalenii_PYR-1 ---------------------
MSMEG_3906|M.smegmatis_MC2_155 ---------------------
TH_0044|M.thermoresistible__bu ---------------------
MAB_2159|M.abscessus_ATCC_1997 ---------------------