For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTTTLARPATRTAVLYRPGSAVFWVFVIALAAGAFALLNQEGSAIHETLDANLALAPFWVAFVAFMVWL LLRFDPFRAMRPYPQVLVAGAVLGATVAVPMAITGNTSLEALWGLVFSPEVVANWGPSLCAPIIEEASKL MCALVLLVLCASVVTRIAHALLIGMFVGLGFDLMEDLGYASSSAILSLDSDLSGAGTSLLVRAFTAVPAH WSYTSLTAVGALLLLPSFAGRRDWPAARRIPVAVGLLLAGPLMHFVWDAPLPDDYDGPLTLVKLVGSLVF YLVIVFALLPFERRWVTARIADGRDSGWLSDVSADVLDSLPTRRQRRALRKAARKSGGRKTAKAVATQQD AALDLIQTAS
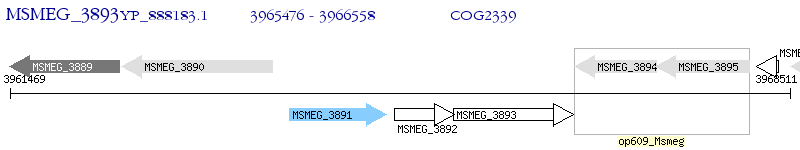
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3893 | - | - | 100% (360) | hypothetical protein MSMEG_3893 |
| M. smegmatis MC2 155 | MSMEG_3838 | - | 7e-16 | 27.53% (356) | integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3562 | - | 2e-06 | 25.52% (337) | hypothetical protein Mflv_3562 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3357 | - | 3e-06 | 26.01% (273) | hypothetical protein Mvan_3357 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3562|M.gilvum_PYR-GCK --MSIAGAPGGPHQFHHHGAPFARRIRTVGAPLGVLIALGTVAGLIVIVL
Mvan_3357|M.vanbaalenii_PYR-1 --MSYAGAPGGPHRFPHYGAPFARRVRSVGAPLGLIIGLGTLAGLIVILL
MSMEG_3893|M.smegmatis_MC2_155 MTTTTLARPATRTAVLYRPGSAVFWVFVIALAAGAFALLNQEG--SAIHE
: . *. . : .. . : :. . * : *. . .*
Mflv_3562|M.gilvum_PYR-GCK TLANPVGTAVGFVLSSVAMAVVVLAYLWLDRWEPEPPRLLIFAFIWGTSV
Mvan_3357|M.vanbaalenii_PYR-1 TLANPVGTAIGFVLSSVAMTLVVLAYLWLDRWEPEPPRLLVFAFVWGTSV
MSMEG_3893|M.smegmatis_MC2_155 TLDANLALAPFWVAFVAFMVWLLLRFDPFRAMRPYP-QVLVAGAVLGATV
** :. * :* . *. ::* : : .* * ::*: . : *::*
Mflv_3562|M.gilvum_PYR-GCK AVVVSAILQVVLDTWLNPGLDPDAADVSAFTLVVGAPMTEEAAKGLFLLL
Mvan_3357|M.vanbaalenii_PYR-1 AVVISAILQIFLDAWLNPAVDPAATGMSPFALVVGAPVTEEAAKGLFLLL
MSMEG_3893|M.smegmatis_MC2_155 AVPMAITGNTSLEALWGLVFSPEVVAN--WGPSLCAPIIEEASKLMCALV
** :: : *:: . ..* .. : : **: ***:* : *:
Mflv_3562|M.gilvum_PYR-GCK MMTGARRRELNSLTDCLVYAGLVGAGFAWLEDILYIANGESLAESLFTAA
Mvan_3357|M.vanbaalenii_PYR-1 MMNGARRNEMNSLTDCLVYAGLVGAGFAWLEDILYIANGESLADSLFTAV
MSMEG_3893|M.smegmatis_MC2_155 LLVLCAS-VVTRIAHALLIGMFVGLGFDLMEDLGYASS---------SAI
:: . :. ::..*: . :** ** :**: * :. :*
Mflv_3562|M.gilvum_PYR-GCK LRLVMGPFAHSLFTTMFALGVWFALHRRTRLGKFGCLLLGYAGAVLLHAM
Mvan_3357|M.vanbaalenii_PYR-1 LRLVMGPFAHSLFTTLFALGVWFALHRRSGIAKAGCILLGYLGAVVLHAM
MSMEG_3893|M.smegmatis_MC2_155 LSLDSDLSGAGTSLLVRAFTAVPAHWSYTSLTAVGALLL--LPSFAGRRD
* * . . . : *: . * : : *.:** :. :
Mflv_3562|M.gilvum_PYR-GCK WNGSSLLGAGAYFGTYVFWMVPVFALTIALAVHSRRREQRIVAAKLPGMV
Mvan_3357|M.vanbaalenii_PYR-1 WNGSSLLGAGAYFGTYVFWMMPVFGLAIALAVDSRRREQRVVAEKLPGMV
MSMEG_3893|M.smegmatis_MC2_155 WPAARRIPVAVGLLLAGPLMHFVWDAPLPDDYDGPLTLVKLVGSLVFYLV
* .: : ... : * *: .:. .. ::*. : :*
Mflv_3562|M.gilvum_PYR-GCK AASVITPSEATWLGSMQTRKYAVAQATRLGGKEAGKSVRRFAHQVTELAF
Mvan_3357|M.vanbaalenii_PYR-1 AAGVITANEATWLGSLRTRRQAVAEATRFGGKPAGDSVKQFAQRVVELAF
MSMEG_3893|M.smegmatis_MC2_155 IVFALLPFERRWV----TARIADGRDSGWLSDVSADVLDSLPTRRQRRAL
. .: . * *: * : * .. : .. :.. : :. : . *:
Mflv_3562|M.gilvum_PYR-GCK VRDRIDRGFGDARVVALLQEETYAVYAARSASPALQQMAGFWTHAPNAPR
Mvan_3357|M.vanbaalenii_PYR-1 VRDRFDRGFGDQRLAALLHEETFAVYAARAASPALQRLAGFRTYAPPAPR
MSMEG_3893|M.smegmatis_MC2_155 RKAARKSG-GRKTAKAVATQQDAALDLIQTAS------------------
: . * * *: :: *: ::**
Mflv_3562|M.gilvum_PYR-GCK P
Mvan_3357|M.vanbaalenii_PYR-1 S
MSMEG_3893|M.smegmatis_MC2_155 -