For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRTQPAPLVFSTSLGLIMAAAVAAPADGPALVAAAISAAAVAVGPAWRPASTVAVWAAAAALALSEPDVL YAALAGLSAAVYLAVRHAAGTSVVTTTRPMMLCALGFTAAAMLVGFWQLTLPWVPLAAPVAVGAVYLIVL RPFVGRARWDVSPAREL*
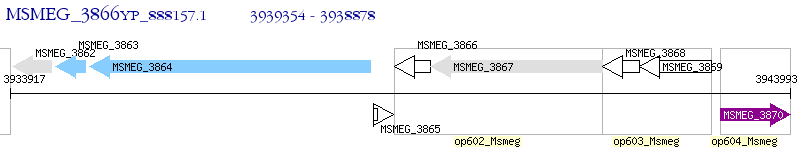
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3866 | - | - | 100% (158) | hypothetical protein MSMEG_3866 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0474 | - | 3e-20 | 41.98% (131) | hypothetical protein Mflv_0474 |
| M. tuberculosis H37Rv | Rv3162c | - | 3e-21 | 39.57% (139) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1401 | - | 1e-25 | 45.65% (138) | hypothetical protein MMAR_1401 |
| M. avium 104 | MAV_4049 | - | 1e-22 | 39.31% (145) | hypothetical protein MAV_4049 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2475 | - | 1e-25 | 45.65% (138) | hypothetical protein MUL_2475 |
| M. vanbaalenii PYR-1 | Mvan_0182 | - | 5e-21 | 44.44% (126) | hypothetical protein Mvan_0182 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1401|M.marinum_M MGS-VAGTGSRVLSPVFGLLMVGAAGAGAQGPVIVALAGAVIALVAGILV
MUL_2475|M.ulcerans_Agy99 MGS-VAGTGSRVLSPVFGLLMVGAAGAGAQGPVIVALAGAVIALVAGILV
Rv3162c|M.tuberculosis_H37Rv MTS-FAHPGTRGLSTVFGLMMVGSAAVGSHGLAVVVGLAAVIAVGVAAVF
MAV_4049|M.avium_104 MDSGRTNLPARVGAPGFGLLMVGLAAGGRPESAAVAVAAALIAVAAGTVL
Mflv_0474|M.gilvum_PYR-GCK MTR-------SLLPVAFGVLMTGAAAYRASGPALLVLGAAVAAVVASAWL
Mvan_0182|M.vanbaalenii_PYR-1 --------------------MVAAAAYRVHGYGVVAAAAALAAVVVSAWW
MSMEG_3866|M.smegmatis_MC2_155 MMR--TQPAPLVFSTSLGLIMAAAVAAPADGPALVAAAISAAAVAVGPAW
*.. .. :. : *: ..
MMAR_1401|M.marinum_M RAAATVAVLLTVLALVLSAPSQLLAALCGLCAVAYLVSRHAAGAPAGVAM
MUL_2475|M.ulcerans_Agy99 RAAATVAVLLTVLALVLSAPPQLLAALCGLCAVAYLVSRHAAGAPAGVAM
Rv3162c|M.tuberculosis_H37Rv RLAATLAVVLSVVMIVVSGPTHVLAALSGFCAAVYLVCRYGAGVVAG---
MAV_4049|M.avium_104 RPAATIAVLLAAVTVVTSNPPPACVALSGLCATAYLVCRHAVGTPVPVIL
Mflv_0474|M.gilvum_PYR-GCK RPAGTVAVVLVVGAIVLTPSAPLHTVLAGLAATVYLVVLHTV--------
Mvan_0182|M.vanbaalenii_PYR-1 RPAATGAVLLAVLTTVLADAAPMHTVLAGLAAAVYLVLRHTN--------
MSMEG_3866|M.smegmatis_MC2_155 RPASTVAVWAAAAALALSEPDVLYAALAGLSAAVYLAVRHAAGTSVVT--
* *.* ** . . : . ..*.*:.*..**. :
MMAR_1401|M.marinum_M ATWPTIVAALGFAFAGLAATAFPLQVPWLPLAAPLAVLAIYVLATRPFLH
MUL_2475|M.ulcerans_Agy99 ATWPTIVAALGFAFAGLAATAFPLQVPWLPLAAPLAVLAIYVLATRPFLH
Rv3162c|M.tuberculosis_H37Rv -SWPTTVAAVGFTFAGLAATSFPLQVPWLPLAAPLAVLATYVLATRPFSR
MAV_4049|M.avium_104 GSRTTAVAAVGFSFAGLVATSFPLQLPWLPLLAPLGALAIYLLAIRPFLS
Mflv_0474|M.gilvum_PYR-GCK ATVPTMVFAVGFGAVAAVVAALPLGVPWLPLIAAPALLAGYLLALRPFTR
Mvan_0182|M.vanbaalenii_PYR-1 PTAPTAFFAVGFAAAAAAVLALPVHVPWLPLAAPLALLAGYLLALKPFTR
MSMEG_3866|M.smegmatis_MC2_155 TTRPMMLCALGFTAAAMLVGFWQLTLPWVPLAAPVAVGAVYLIVLRPFVG
: . . *:** .. . : :**:** *. . * *::. :**
MMAR_1401|M.marinum_M ------------
MUL_2475|M.ulcerans_Agy99 ------------
Rv3162c|M.tuberculosis_H37Rv ------------
MAV_4049|M.avium_104 ------------
Mflv_0474|M.gilvum_PYR-GCK ------------
Mvan_0182|M.vanbaalenii_PYR-1 ------------
MSMEG_3866|M.smegmatis_MC2_155 RARWDVSPAREL