For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AKRLFLVYALVELAVMVVLVATIGFGWTVLLLLAAFVVGLALAGSQARRQLRKLSGGLTAASAQGAVTDS LLVALGTVLVVVPGLASSVLGAVLLLPPTRAAARPVLTAMAAKRVPLVTGFQTASRYQWTAGYPGGRGDY IDGEVIDVTDSADAAGGVEPHRLPPKA*
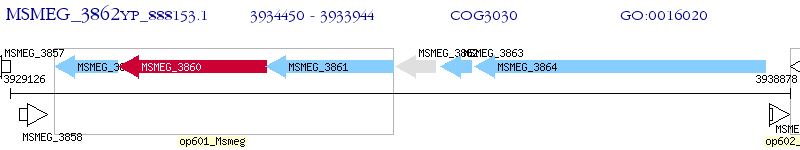
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3862 | fxsA | - | 100% (168) | FxsA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2079c | fxsA | 9e-30 | 47.37% (171) | FxsA |
| M. gilvum PYR-GCK | Mflv_3115 | fxsA | 5e-53 | 64.24% (165) | FxsA |
| M. tuberculosis H37Rv | Rv2053c | fxsA | 1e-29 | 47.37% (171) | FxsA |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2204 | fxsA | 1e-35 | 50.30% (167) | FxsA |
| M. marinum M | MMAR_3031 | fxsA | 2e-28 | 45.09% (173) | hypothetical protein MMAR_3031 |
| M. avium 104 | MAV_2444 | fxsA | 2e-26 | 44.87% (156) | FxsA |
| M. thermoresistible (build 8) | TH_2630 | - | 4e-42 | 54.26% (188) | PUTATIVE putative FxsA cytoplasmic membrane protein |
| M. ulcerans Agy99 | MUL_2271 | fxsA | 6e-29 | 45.09% (173) | FxsA |
| M. vanbaalenii PYR-1 | Mvan_3421 | fxsA | 2e-51 | 68.92% (148) | FxsA |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3115|M.gilvum_PYR-GCK MAMRLFLLYAVIELAVLVALVSTIGFGWTLLLVAGAFVLGL----ALAGS
Mvan_3421|M.vanbaalenii_PYR-1 --MRLFLIYVVVEMAVVVALASTIGFGWTVLLLAGAFLLGL----ALAGS
MSMEG_3862|M.smegmatis_MC2_155 MAKRLFLVYALVELAVMVVLVATIGFGWTVLLLLAAFVVGL----ALAGS
TH_2630|M.thermoresistible__bu MAQRLFLLYAVVELAVFIAMVTTVGFGWAVLAVLVAFLLGL----VLAGA
MAB_2204|M.abscessus_ATCC_1997 MWPGLFLLYVIVEVSALVALTSTVGIGWTILAVAGAFVIGL----VLAGS
Mb2079c|M.bovis_AF2122/97 -MSRLLLSYAVVELAVVFALAATIGFGWTLLVLLATFVLGFGLLAPLGGW
Rv2053c|M.tuberculosis_H37Rv -MSRLLLSYAVVELAVVFALAATIGFGWTLLVLLATFVLGFGLLAPLGGW
MMAR_3031|M.marinum_M MVGRLILIYAVVELMALIGLTAAIGFPWAVLVLLASFVLGLVLWAPMGGW
MUL_2271|M.ulcerans_Agy99 MVGRLILIYAVVELMALIGLTAAIGFAWAVLVLLASFVLGLVLWAPMGGW
MAV_2444|M.avium_104 MVARLLLLYAVVELAVTFAVVSTIGWGSTLLVLLATFLLGWGVVAPMAGS
*:* *.::*: . . :.:::* ::* : :*::* :.*
Mflv_3115|M.gilvum_PYR-GCK QLRRHLRRLRSGLTLADAQG-------AVTDSVLVALGTVLVVIPGLASS
Mvan_3421|M.vanbaalenii_PYR-1 QLRRHLRRLQSGLTLADAQG-------AVTDSVLVALGTVLVVIPGLASS
MSMEG_3862|M.smegmatis_MC2_155 QARRQLRKLSGGLTAASAQG-------AVTDSLLVALGTVLVVVPGLASS
TH_2630|M.thermoresistible__bu QVTRQLRELRSTLAGAAPPGRLGSPLGAATDSLMVAAGTVLVVVPGLASS
MAB_2204|M.abscessus_ATCC_1997 QARRALGQLRRG--GRSPGG-------AIADGALIALGTIAVVIPGLVSS
Mb2079c|M.bovis_AF2122/97 QLGRRLLWLRSGLAEPRS---------ALSDGALVTVASVLVLVPGLVTT
Rv2053c|M.tuberculosis_H37Rv QLGRRLLWLRSGLAEPRS---------ALSDGALVTVASVLVLVPGLVTT
MMAR_3031|M.marinum_M QLSRQLGQLRSGLQEPRS---------VLSDGALVAVATGLVLVPGLVTT
MUL_2271|M.ulcerans_Agy99 QLSRQLGQLRSGLQEPRS---------VLSDGALVAVATGLVLVPGLVTT
MAV_2444|M.avium_104 QLLRRIGQLRSGLTEPRA---------AAGDGALLSVATALVLIPGLLST
* * : * . . *. ::: .: *::*** ::
Mflv_3115|M.gilvum_PYR-GCK VFGALLLLPPTRAAARPLVTALAARRTPLIVGVSTVG----SAATRRAGR
Mvan_3421|M.vanbaalenii_PYR-1 VLGALLLLPPTRAAARPMITAMAARRAPLIVGVTTVG----SAAG-RARR
MSMEG_3862|M.smegmatis_MC2_155 VLGAVLLLPPTRAAARPVLTAMAAKRVPLVTGFQTASRY-QWTAGYPGGR
TH_2630|M.thermoresistible__bu VLGALLLAPPTRAVLRPAITAVAARRISRHLPLITVA-------GHRAGR
MAB_2204|M.abscessus_ATCC_1997 ALGLLLLLPPTRAVLRPLLAFVAARQLSRRAPLIGVIPTGYGAYQRTAPR
Mb2079c|M.bovis_AF2122/97 TMGLLLLVPPIRALARPGLTAIAVRGFLRNVPLTADAAANMAGAFG--ES
Rv2053c|M.tuberculosis_H37Rv TMGLLLLVPPIRALARPGLTAIAVRGFLRNVPLTADAAANMAGAFG--ES
MMAR_3031|M.marinum_M VFGLLLLAPPIRAAARPGLTAIAVRGLLRRVPLTGAAAASMADAFNRRNA
MUL_2271|M.ulcerans_Agy99 VFGLLLLAPPIRAAARPGLTAIAVRGLLRRVPLTGAAAASMADAFNRRNA
MAV_2444|M.avium_104 ALGLMLFIPPVRGAAAPRLTALTLRTLQRHAPPYPTAFRAATDPRG---P
.:* :*: ** *. * :: :: :
Mflv_3115|M.gilvum_PYR-GCK A---EYIDGEVVE------VIDDVPVHDGGVPSPRPEL------------
Mvan_3421|M.vanbaalenii_PYR-1 R---DYIDGEVVE------VVD--------VPPVSTDQ------------
MSMEG_3862|M.smegmatis_MC2_155 G---DYIDGEVID------VTDS------ADAAGGVEP------------
TH_2630|M.thermoresistible__bu P---GYIDGEVIDAHVVEHVTDTVTDTDVAGTVIDADQ------------
MAB_2204|M.abscessus_ATCC_1997 PTTVDYIDGEVID------VADE----SGPQTYRYRPG------------
Mb2079c|M.bovis_AF2122/97 GTDPDFIDGEVID------VIDVEP-LTLQPPRVAAEP------------
Rv2053c|M.tuberculosis_H37Rv GTDPDFIDGEVID------VIDVEP-LTLQPPRVAAEP------------
MMAR_3031|M.marinum_M REQRDFIDGEVID------VIDGEP-PTLPHAVVEVDELDEFDDFDGFDR
MUL_2271|M.ulcerans_Agy99 REQRDFIDGEVID------VIDGEP-PTLPQAVVEVDELDEFDDFDGFDR
MAV_2444|M.avium_104 YEQRDYIDGEVID------VHDVPA-PARNQPFDG---------------
:*****:: * * .
Mflv_3115|M.gilvum_PYR-GCK -------------------PRTPE--
Mvan_3421|M.vanbaalenii_PYR-1 -------------------PRLPE--
MSMEG_3862|M.smegmatis_MC2_155 -------------------HRLPPKA
TH_2630|M.thermoresistible__bu -------------------PRLPRTD
MAB_2204|M.abscessus_ATCC_1997 -------------------HDLPA--
Mb2079c|M.bovis_AF2122/97 --------------------PSPGSN
Rv2053c|M.tuberculosis_H37Rv --------------------PSPGSN
MMAR_3031|M.marinum_M FNGFTRFDSFGNVRTSTRHRSAPGAR
MUL_2271|M.ulcerans_Agy99 FNGFTRFDSFGNVRTSTRHRSAPGAR
MAV_2444|M.avium_104 -------------------RRATG--
.