For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSPSVTSPQVAVNDIGSAEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDEVLLDIGYKTEGVIPSRELSIK HDVDPNEVVSVGDEVEALVLTKEDKEGRLILSKKRAQYERAWGTIEELKEKDEAVKGTVIEVVKGGLILD IGLRGFLPASLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRSEFLNQLQKGAI RKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSEVVQVGDEVTVEVLDVDMDRERVSLSLKATQE DPWRHFARTHAIGQIVPGKVTKLVPFGAFVRVEEGIEGLVHISELSERHVEVPDQVVQVGDDAMVKVIDI DLERRRISLSLKQANEDYTEEFDPSKYGMADSYDEQGNYIFPEGFDPETNEWLEGFDKQREEWEARYAEA ERRHKMHTAQMEKFAAAEAEAANAPVSNGSSRSEESSGGTLASDAQLAALREKLAGNA*
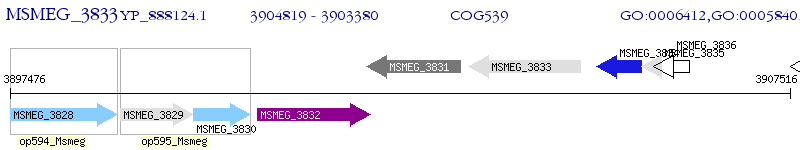
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3833 | rpsA | - | 100% (479) | 30S ribosomal protein S1 |
| M. smegmatis MC2 155 | MSMEG_2839 | - | 2e-14 | 47.56% (82) | transcriptional accessory protein |
| M. smegmatis MC2 155 | MSMEG_2656 | gpsI | 2e-07 | 46.38% (69) | polynucleotide phosphorylase/polyadenylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1656 | rpsA | 0.0 | 93.76% (481) | 30S ribosomal protein S1 |
| M. gilvum PYR-GCK | Mflv_3561 | rpsA | 0.0 | 96.47% (481) | 30S ribosomal protein S1 |
| M. tuberculosis H37Rv | Rv1630 | rpsA | 0.0 | 93.97% (481) | 30S ribosomal protein S1 |
| M. leprae Br4923 | MLBr_01382 | rpsA | 0.0 | 91.06% (481) | 30S ribosomal protein S1 |
| M. abscessus ATCC 19977 | MAB_2296 | rpsA | 0.0 | 95.23% (482) | 30S ribosomal protein S1 |
| M. marinum M | MMAR_2433 | rpsA | 0.0 | 93.96% (480) | ribosomal protein S1 RpsA |
| M. avium 104 | MAV_3152 | rpsA | 0.0 | 94.79% (480) | 30S ribosomal protein S1 |
| M. thermoresistible (build 8) | TH_0575 | rpsA | 0.0 | 94.99% (479) | PROBABLE RIBOSOMAL PROTEIN S1 RPSA |
| M. ulcerans Agy99 | MUL_1612 | rpsA | 0.0 | 93.96% (480) | 30S ribosomal protein S1 |
| M. vanbaalenii PYR-1 | Mvan_3356 | rpsA | 0.0 | 95.63% (481) | 30S ribosomal protein S1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1656|M.bovis_AF2122/97 MPSPTVTSPQVAVNDIGSSEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDE
Rv1630|M.tuberculosis_H37Rv MPSPTVTSPQVAVNDIGSSEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDE
Mflv_3561|M.gilvum_PYR-GCK MPSPSVTSPQVAVNDIGSAEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDE
Mvan_3356|M.vanbaalenii_PYR-1 MPSPSVTSPQVAVNDIGSAEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDE
TH_0575|M.thermoresistible__bu MPSPSVTSPQVAVNDIGSSEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDE
MSMEG_3833|M.smegmatis_MC2_155 MPSPSVTSPQVAVNDIGSAEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDE
MAB_2296|M.abscessus_ATCC_1997 MPSPTVTSPQVAVNDIGSAEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDE
MMAR_2433|M.marinum_M MPSPTVTSPQVAVNDIGSAEDFLAAIDQTIKYFNDGDIVEGTIVKVDRDE
MUL_1612|M.ulcerans_Agy99 MPSPTVTSPQVAVNDIGSAEDFLAAIDQTIKYFNDGDIVEGTIVKVDRDE
MAV_3152|M.avium_104 MPSPAVTSPQVAVNDIGSSEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDE
MLBr_01382|M.leprae_Br4923 MSIPAVPSPQIAVNDVGSSEDFLAAIDKTIKYFNDGDIVEGTIVKVDRDE
*. *:*.***:****:**:********:**********************
Mb1656|M.bovis_AF2122/97 VLLDIGYKTEGVIPARELSIKHDVDPNEVVSVGDEVEALVLTKEDKEGRL
Rv1630|M.tuberculosis_H37Rv VLLDIGYKTEGVIPARELSIKHDVDPNEVVSVGDEVEALVLTKEDKEGRL
Mflv_3561|M.gilvum_PYR-GCK VLLDIGYKTEGVIPSRELSIKHDVDPNEVVSVGDEVEALVLTKEDKEGRL
Mvan_3356|M.vanbaalenii_PYR-1 VLLDIGYKTEGVIPSRELSIKHDVDPNEVVSVGDEVEALVLTKEDKEGRL
TH_0575|M.thermoresistible__bu VLLDIGYKTEGVIPSRELSIKHDVDPHEVVSVGDQVEALVLTKEDKDGRL
MSMEG_3833|M.smegmatis_MC2_155 VLLDIGYKTEGVIPSRELSIKHDVDPNEVVSVGDEVEALVLTKEDKEGRL
MAB_2296|M.abscessus_ATCC_1997 VLLDIGYKTEGVIPSRELSIKHDVDPNEVVSVGDEVEALVLTKEDKEGRL
MMAR_2433|M.marinum_M VLLDIGYKTEGVIPARELSIKHDVDPNEVVTVGDEVEALVLTKEDKEGRL
MUL_1612|M.ulcerans_Agy99 VLLDIGYKTEGVIPARELSIKHDVDPNEVVTVGDEVEALVLTKEDKEGRL
MAV_3152|M.avium_104 VLLDIGYKTEGVIPARELSIKHDVDPNEVVSVGDEVEALVLTKEDKEGRL
MLBr_01382|M.leprae_Br4923 VLLDIGYKTEGVIPARELSIKHDVDPNEVVSVGDEVEALVLTKEDKEGRL
**************:***********:***:***:***********:***
Mb1656|M.bovis_AF2122/97 ILSKKRAQYERAWGTIEALKEKDEAVKGTVIEVVKGGLILDIGLRGFLPA
Rv1630|M.tuberculosis_H37Rv ILSKKRAQYERAWGTIEALKEKDEAVKGTVIEVVKGGLILDIGLRGFLPA
Mflv_3561|M.gilvum_PYR-GCK ILSKKRAQYERAWGTIEELKEKDEAVKGTVIEVVKGGLILDIGLRGFLPA
Mvan_3356|M.vanbaalenii_PYR-1 ILSKKRAQYERAWGTIEELKEKDEAVKGTVIEVVKGGLILDIGLRGFLPA
TH_0575|M.thermoresistible__bu ILSKKRAQYERAWGTIEALKEKDEPVKGTVIEVVKGGLILDIGLRGFLPA
MSMEG_3833|M.smegmatis_MC2_155 ILSKKRAQYERAWGTIEELKEKDEAVKGTVIEVVKGGLILDIGLRGFLPA
MAB_2296|M.abscessus_ATCC_1997 ILSKKRAQYERAWGTIEELKEKDEAVKGTVIEVVKGGLILDIGLRGFLPA
MMAR_2433|M.marinum_M ILSKKRAQYERAWGTIEALKEKDEAVKGTVIEVVKGGLILDIGLRGFLPA
MUL_1612|M.ulcerans_Agy99 ILSKKRAQYERAWGTIEALKEKDEAVKGTVIEVVKGGLILDIGLRGFLPA
MAV_3152|M.avium_104 ILSKKRAQYERAWGTIEALKEKDEAVKGTVIEVVKGGLILDIGLRGFLPA
MLBr_01382|M.leprae_Br4923 ILSKKRAQYERAWGTIEALKEKDEAVKGIVIEVVKGGLILDIGLRGFLPA
***************** ******.*** *********************
Mb1656|M.bovis_AF2122/97 SLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
Rv1630|M.tuberculosis_H37Rv SLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
Mflv_3561|M.gilvum_PYR-GCK SLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
Mvan_3356|M.vanbaalenii_PYR-1 SLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
TH_0575|M.thermoresistible__bu SLVEMRRVRDLQPYIGREIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
MSMEG_3833|M.smegmatis_MC2_155 SLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
MAB_2296|M.abscessus_ATCC_1997 SLVEMRRVRDLQPYIGKEIEAKIIELDRNRNNVVLSRRAWLEQTQSEVRS
MMAR_2433|M.marinum_M SLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
MUL_1612|M.ulcerans_Agy99 SLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
MAV_3152|M.avium_104 SLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
MLBr_01382|M.leprae_Br4923 SLVEMRRVRDLQPYIGKEIEAKIIELDKNRNNVVLSRRAWLEQTQSEVRS
****************:**********:**********************
Mb1656|M.bovis_AF2122/97 EFLNNLQKGTIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
Rv1630|M.tuberculosis_H37Rv EFLNNLQKGTIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
Mflv_3561|M.gilvum_PYR-GCK EFLNQLTKGAIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
Mvan_3356|M.vanbaalenii_PYR-1 EFLNQLTKGAIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
TH_0575|M.thermoresistible__bu EFLNQLQKGAIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
MSMEG_3833|M.smegmatis_MC2_155 EFLNQLQKGAIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
MAB_2296|M.abscessus_ATCC_1997 EFLNQLTKGAIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
MMAR_2433|M.marinum_M EFLNQLQKGTIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
MUL_1612|M.ulcerans_Agy99 EFLNQLQKGTIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
MAV_3152|M.avium_104 EFLNQLQKGAIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
MLBr_01382|M.leprae_Br4923 EFLNQLQKGAIRKGVVSSIVNFGAFVDLGGVDGLVHVSELSWKHIDHPSE
****:* **:****************************************
Mb1656|M.bovis_AF2122/97 VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
Rv1630|M.tuberculosis_H37Rv VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
Mflv_3561|M.gilvum_PYR-GCK VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
Mvan_3356|M.vanbaalenii_PYR-1 VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
TH_0575|M.thermoresistible__bu VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
MSMEG_3833|M.smegmatis_MC2_155 VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
MAB_2296|M.abscessus_ATCC_1997 VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
MMAR_2433|M.marinum_M VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
MUL_1612|M.ulcerans_Agy99 VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
MAV_3152|M.avium_104 VVQVGDEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
MLBr_01382|M.leprae_Br4923 VVQVGNEVTVEVLDVDMDRERVSLSLKATQEDPWRHFARTHAIGQIVPGK
*****:********************************************
Mb1656|M.bovis_AF2122/97 VTKLVPFGAFVRVEEGIEGLVHISELAERHVEVPDQVVAVGDDAMVKVID
Rv1630|M.tuberculosis_H37Rv VTKLVPFGAFVRVEEGIEGLVHISELAERHVEVPDQVVAVGDDAMVKVID
Mflv_3561|M.gilvum_PYR-GCK VTKLVPFGAFVRVEEGIEGLVHISELSERHVEVPDQVVQVGDDAMVKVID
Mvan_3356|M.vanbaalenii_PYR-1 VTKLVPFGAFVRVEEGIEGLVHISELSERHVEVPDQVVQVGDDAMVKVID
TH_0575|M.thermoresistible__bu VTKLVPFGAFVRVEEGIEGLVHISELSERHIEVPDQVVQVGDDVMVKVID
MSMEG_3833|M.smegmatis_MC2_155 VTKLVPFGAFVRVEEGIEGLVHISELSERHVEVPDQVVQVGDDAMVKVID
MAB_2296|M.abscessus_ATCC_1997 VTKLVPFGAFVRVEEGIEGLVHISELSERHVEVPDQVVGVGDDAMVKVID
MMAR_2433|M.marinum_M VTKLVPFGAFVRVEEGIEGLVHISELAERHVEVPDQVVAVGDDAMVKVID
MUL_1612|M.ulcerans_Agy99 VTKLVPFGAFVRVEEGIEGLVHISELAERHVEVPDQVVAVGDDAMVKVID
MAV_3152|M.avium_104 VTKLVPFGAFVRVEEGIEGLVHISELAERHVEVPDQVVAVGDDAMVKVID
MLBr_01382|M.leprae_Br4923 VTKLVPFGAFVRVEEGIEGLVHISELAERHVEVPDQVVAVGDDAMVKVID
**************************:***:******* ****.******
Mb1656|M.bovis_AF2122/97 IDLERRRISLSLKQANEDYTEEFDPAKYGMADSYDEQGNYIFPEGFDAET
Rv1630|M.tuberculosis_H37Rv IDLERRRISLSLKQANEDYTEEFDPAKYGMADSYDEQGNYIFPEGFDAET
Mflv_3561|M.gilvum_PYR-GCK IDLERRRISLSLKQANEDYSEEFEAWKYGMADSYDDQGNYIFPEGFDADT
Mvan_3356|M.vanbaalenii_PYR-1 IDLERRRISLSLKQANEDYSEEFEAWKYGMADSYDDQGNYIFPEGFDADT
TH_0575|M.thermoresistible__bu IDLDRRRISLSLKQANEDYSEEFDPSKYGMADSYDEQGNYIFPEGFDPET
MSMEG_3833|M.smegmatis_MC2_155 IDLERRRISLSLKQANEDYTEEFDPSKYGMADSYDEQGNYIFPEGFDPET
MAB_2296|M.abscessus_ATCC_1997 IDLERRRISLSLKQANEDYTEEFDPSKYGMADSYDDAGNYIFPEGFDAET
MMAR_2433|M.marinum_M IDLERRRISLSLKQANEDYTDEFDPAKYGMADSYDDQGNYIFPEGFDPET
MUL_1612|M.ulcerans_Agy99 IDLERRRISLSLKQANEDYTDEFDPAKYGMADSYDDQGNYIFPEGFDPET
MAV_3152|M.avium_104 IDLERRRISLSLKQANEDYTEEFDPAKYGMADSYDEQGNYIFPEGFDPET
MLBr_01382|M.leprae_Br4923 IDLERRRISLSLKQANEDYIEEFDPAKYGMADSYDEQGNYIFPEGFDPDS
***:*************** :**:. *********: **********.::
Mb1656|M.bovis_AF2122/97 NEWLEGFEKQRAEWEARYAEAERRHKMHTAQMEKFAAAETAGRGADDQSS
Rv1630|M.tuberculosis_H37Rv NEWLEGFEKQRAEWEARYAEAERRHKMHTAQMEKFAAAEAAGRGADDQSS
Mflv_3561|M.gilvum_PYR-GCK NEWLEGFEKQRDEWEARYAEAERRHKMHTAQMEKFAAAEAEEAAKPASSN
Mvan_3356|M.vanbaalenii_PYR-1 NEWLEGFEKQRDEWEARYAEAERRHKMHTAQMEKFAAAEAEEAARPTSSS
TH_0575|M.thermoresistible__bu NEWLEGFEKQREEWEARYAEAERRHKMHTAQMEKFAAAEAEAASRPATST
MSMEG_3833|M.smegmatis_MC2_155 NEWLEGFDKQREEWEARYAEAERRHKMHTAQMEKFAAAEAEAANAPVSNG
MAB_2296|M.abscessus_ATCC_1997 NEWLEGFDKQREEWEARYAEAERRHKMHTVQMEKSAAAAEAEAAAASTSS
MMAR_2433|M.marinum_M NEWMEGFDTQRAEWEARYAEAERRHKMHTTQMEKFAAAEAAGHGDNGGGS
MUL_1612|M.ulcerans_Agy99 NEWMEGFDTQRAEWEARYAEAERRHKMHTTQMEKFAAAEAAGHGDNGGGS
MAV_3152|M.avium_104 NEWLEGFEKQRAEWEARYAEAERRHKMHTAQMEKFAAAEAAGHAGGEQSP
MLBr_01382|M.leprae_Br4923 NEWLEGFDTQRAEWEARYAEAERRYKMHTIQMEKFAATEEAGHGSSEQPP
***:***:.** ************:**** **** **:
Mb1656|M.bovis_AF2122/97 ASSAPSEKTAGGSLASDAQLAALREKLAGSA
Rv1630|M.tuberculosis_H37Rv ASSAPSEKTAGGSLASDAQLAALREKLAGSA
Mflv_3561|M.gilvum_PYR-GCK GSSRSEESSGGGSLASDAQLAALREKLAGNA
Mvan_3356|M.vanbaalenii_PYR-1 SNGARSEESAGGSLASDAQLAALREKLAGNA
TH_0575|M.thermoresistible__bu S---RSDEPAGGSLASDEQLAALREKLAGNA
MSMEG_3833|M.smegmatis_MC2_155 SS--RSEESSGGTLASDAQLAALREKLAGNA
MAB_2296|M.abscessus_ATCC_1997 SSRSDDSASQGGSLANDEQLAALREKLAGNA
MMAR_2433|M.marinum_M SSSSSSEKSAGGSLASDAQLAALREKLAGNG
MUL_1612|M.ulcerans_Agy99 SSSSSSEKSAGGSLASDAQLAALREKLAGNG
MAV_3152|M.avium_104 GNGAPAEK-AGGSLASDAQLAALREKLAGNA
MLBr_01382|M.leprae_Br4923 ASSTPSAKATGGSLASDAQLAALREKLAGSA
. **:**.* ***********..