For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PALRKHPQTATKHLFVTGGVVSSLGKGLTASSLGQLLTARGLQVTMQKLDPYLNVDPGTMNPFQHGEVFV TEDGAETDLDVGHYERFLDRNLSGSANVTTGQVYSSVIAKERRGEYLGDTVQVIPHITDEIKSRILAMAE PDAAGVRPDVVITEVGGTVGDIESLPFLEAARQVRHEVGRENCFFLHVSLVPYLAPSGELKTKPTQHSVA ALRSIGITPDALILRCDRDVPEPLKNKIALMCDVDVDGVISTPDAPSIYDIPKVLHREELDAYVVRRLNL PFRDVDWTEWDDLLRRVHEPQETVRIALVGKYIDLSDAYLSVAEALRAGGFKHRAKVEMRWVASDDCETE HGAAAALSDVHGVLIPGGFGIRGIEGKIGAISYARKRGLPVLGLCLGLQCIVIEAARSVGITEANSAEFD PKTPDPVISTMADQRDAVAGEADLGGTMRLGAYPAVLTPNSVVAQAYQSTEVSERHRHRFEVNNAYRDRI AKSGLRFSGTSPDGHLVEFVEYDPQIHPFLVGTQAHPELKSRPTRPHPLFAAFIGAAIDYKAAERLPGMD LPEQFVPVEHSDADAPALEEPLEKSDVRG*
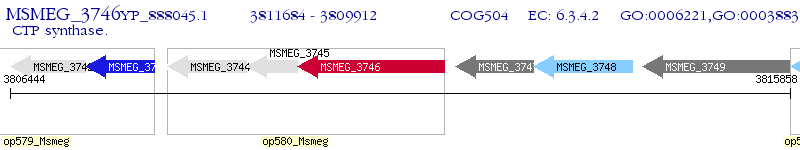
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3746 | pyrG | - | 100% (590) | CTP synthetase |
| M. smegmatis MC2 155 | MSMEG_2596 | - | 8e-05 | 29.67% (182) | peptidase C26 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1725 | pyrG | 0.0 | 86.37% (587) | CTP synthetase |
| M. gilvum PYR-GCK | Mflv_3502 | pyrG | 0.0 | 87.50% (584) | CTP synthetase |
| M. tuberculosis H37Rv | Rv1699 | pyrG | 0.0 | 86.37% (587) | CTP synthetase |
| M. leprae Br4923 | MLBr_01363 | pyrG | 0.0 | 84.39% (583) | CTP synthetase |
| M. abscessus ATCC 19977 | MAB_2364 | pyrG | 0.0 | 87.46% (558) | CTP synthetase |
| M. marinum M | MMAR_2504 | pyrG | 0.0 | 85.59% (583) | CTP synthase, PyrG |
| M. avium 104 | MAV_3072 | pyrG | 0.0 | 86.72% (580) | CTP synthetase |
| M. thermoresistible (build 8) | TH_1544 | pyrG | 0.0 | 85.44% (577) | Probable CTP synthase PyrG |
| M. ulcerans Agy99 | MUL_1688 | pyrG | 0.0 | 85.25% (583) | CTP synthetase |
| M. vanbaalenii PYR-1 | Mvan_3283 | pyrG | 0.0 | 89.73% (584) | CTP synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1725|M.bovis_AF2122/97 -------------MRKHPQTATKHLFVSGGVASSLGKGLTASSLGQLLTA
Rv1699|M.tuberculosis_H37Rv -------------MRKHPQTATKHLFVSGGVASSLGKGLTASSLGQLLTA
MMAR_2504|M.marinum_M -------------MRRHPQTATKHLFVSGGVASSLGKGLTASSLGQLLTA
MUL_1688|M.ulcerans_Agy99 -------------MRRHPQTATKHLFVSGGVASSLGKGLTASSLGQLLTA
MLBr_01363|M.leprae_Br4923 -------------MRKHPQSATKHLFVSGGVASSLGKGLTASSLGQLLTA
MAV_3072|M.avium_104 -------------MRKHPQTATKHLFVSGGVASSLGKGLTASSLGQLLTA
MAB_2364|M.abscessus_ATCC_1997 MPCAVVTGANLAGLRKHPQTATKHLFVSGGVASSLGKGLTASSLGQLLTS
Mflv_3502|M.gilvum_PYR-GCK ----------MPALRKHPHTATKHLFVTGGVVSSLGKGLTASSLGQLLTA
Mvan_3283|M.vanbaalenii_PYR-1 ----------MPALRKHPQTATKHLFVTGGVVSSLGKGLTASSLGQLLTA
MSMEG_3746|M.smegmatis_MC2_155 ----------MPALRKHPQTATKHLFVTGGVVSSLGKGLTASSLGQLLTA
TH_1544|M.thermoresistible__bu ------------------------------VVSSLGKGLTGASLGQLLTA
*.********.:*******:
Mb1725|M.bovis_AF2122/97 RGLHVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
Rv1699|M.tuberculosis_H37Rv RGLHVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
MMAR_2504|M.marinum_M RGLRVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
MUL_1688|M.ulcerans_Agy99 RGLRVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
MLBr_01363|M.leprae_Br4923 RGLHVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
MAV_3072|M.avium_104 RGLYVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
MAB_2364|M.abscessus_ATCC_1997 RGLQVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
Mflv_3502|M.gilvum_PYR-GCK RGLQVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
Mvan_3283|M.vanbaalenii_PYR-1 RGLQVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
MSMEG_3746|M.smegmatis_MC2_155 RGLQVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDVGHYERFLD
TH_1544|M.thermoresistible__bu RGLYVTMQKLDPYLNVDPGTMNPFQHGEVFVTEDGAETDLDIGHYERFLD
*** *************************************:********
Mb1725|M.bovis_AF2122/97 RNLPGSANVTTGQVYSTVIAKERRGEYLGDTVQVIPHITDEIKRRILAMA
Rv1699|M.tuberculosis_H37Rv RNLPGSANVTTGQVYSTVIAKERRGEYLGDTVQVIPHITDEIKRRILAMA
MMAR_2504|M.marinum_M RDLSGSANVTTGQVYSTVIAKERRGEYLGDTVQVIPHITDEIKRRIMAMA
MUL_1688|M.ulcerans_Agy99 RDLSGSANVTTGQVYSTVIAKERRGEYLGDTVQVIPHITDEIKRRIMAMA
MLBr_01363|M.leprae_Br4923 RDLSGSANVTTGQVYSTVIAKERRGEYLGDTVQVIPHITDEIKQRIMAMA
MAV_3072|M.avium_104 RDLSGSANVTTGQVYSTVIAKERRGEYLGDTVQVIPHITDEIKGRILAMA
MAB_2364|M.abscessus_ATCC_1997 RDLSGSANVTTGQVYSSVIAKERRGEYLGDTVQVIPHITDEIKSRILAMA
Mflv_3502|M.gilvum_PYR-GCK RNLSGSANVTTGQVYSTVIAKERRGEYLGDTVQVIPHITDEIKRRVLEMA
Mvan_3283|M.vanbaalenii_PYR-1 RNLSGSANVTTGQVYSTVIAKERRGEYLGDTVQVIPHITDEIKRRILAMD
MSMEG_3746|M.smegmatis_MC2_155 RNLSGSANVTTGQVYSSVIAKERRGEYLGDTVQVIPHITDEIKSRILAMA
TH_1544|M.thermoresistible__bu RDLPGSANVTTGQVYSTVIAKERRGEYLGDTVQVIPHITDEIKRRIMAMA
*:*.************:************************** *:: *
Mb1725|M.bovis_AF2122/97 QPDADGNRPDVVITEIGGTVGDIESQPFLEAARQVRHYLGREDVFFLHVS
Rv1699|M.tuberculosis_H37Rv QPDADGNRPDVVITEIGGTVGDIESQPFLEAARQVRHYLGREDVFFLHVS
MMAR_2504|M.marinum_M EPNADGRRPDVVITEIGGTVGDIESQPFLEAARQVRHDLGRENVFFLHVS
MUL_1688|M.ulcerans_Agy99 EPNADGRRPDVVITEIGGTVGDIESQPFLEAARQVRHDLGRENVFFLHVS
MLBr_01363|M.leprae_Br4923 QPDGGDNRPDVVITEIGGTVGDIESQPFLEAARQVRHDLGRENVFFLHVS
MAV_3072|M.avium_104 QPDAQGNRPDVVITEIGGTVGDIESQPFLEAARQVRHDLGRENVFFLHVS
MAB_2364|M.abscessus_ATCC_1997 EPNENGQRPDIVITEIGGTVGDIESQPFLEAARQIRHDVGRDNVFFLHVS
Mflv_3502|M.gilvum_PYR-GCK EPDEDGNRPDVVITEVGGTVGDIESLPFLEAARQVRHEVGRENCFFLHCS
Mvan_3283|M.vanbaalenii_PYR-1 AADSDGNRPDVIITEIGGTVGDIESLPFLEAARQVRHEVGRENCFFLHCS
MSMEG_3746|M.smegmatis_MC2_155 EPDAAGVRPDVVITEVGGTVGDIESLPFLEAARQVRHEVGRENCFFLHVS
TH_1544|M.thermoresistible__bu EPDADGNRPDVIITEIGGTVGDIESLPFLEAARQVRHEVGRENCFFLHVS
.: . ***::***:********* ********:** :**:: **** *
Mb1725|M.bovis_AF2122/97 LVPYLAPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPEALKNKIA
Rv1699|M.tuberculosis_H37Rv LVPYLAPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPEALKNKIA
MMAR_2504|M.marinum_M LVPYLAPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPEALKNKIA
MUL_1688|M.ulcerans_Agy99 LVPYLAPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPEALKNKIA
MLBr_01363|M.leprae_Br4923 LVPHLAPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPESLKNKIA
MAV_3072|M.avium_104 LVPYLAPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPEALKNKIA
MAB_2364|M.abscessus_ATCC_1997 LVPYLKPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPEPLKNKIA
Mflv_3502|M.gilvum_PYR-GCK LVPFMAPSGELKTKPTQHSVAALRSIGIQPDALILRCDRDVPESLKNKIA
Mvan_3283|M.vanbaalenii_PYR-1 LVPYMAPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPEPLKNKIA
MSMEG_3746|M.smegmatis_MC2_155 LVPYLAPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPEPLKNKIA
TH_1544|M.thermoresistible__bu LVPYLAPSGELKTKPTQHSVAALRSIGITPDALILRCDRDVPDSLKNKIA
***.: ********************** *************:.******
Mb1725|M.bovis_AF2122/97 LMCDVDIDGVISTPDAPSIYDIPKVLHREELDAFVVRRLNLPFRDVDWTE
Rv1699|M.tuberculosis_H37Rv LMCDVDIDGVISTPDAPSIYDIPKVLHREELDAFVVRRLNLPFRDVDWTE
MMAR_2504|M.marinum_M LMCDVDIDGIISTPDAPSIYDIPKVLHREELDAFVVRRLNLPFRDVDWTE
MUL_1688|M.ulcerans_Agy99 LMCDVDIDGIISTPAAPSIYDIPKVLHREELDAFVVRRLNLPFRDVDWTE
MLBr_01363|M.leprae_Br4923 LMCDVDIDGVISTPDAPSIYDIPKVLHREELDAFVVRRLNLPFRDVDWTE
MAV_3072|M.avium_104 LMCDVDIDGVISTPDAPSIYDIPKVLHREELDAYVVRRLNLPFRDVDWTE
MAB_2364|M.abscessus_ATCC_1997 LMCDVDVDGVISTPDAPSIYDIPKVLHREELDAYVVRRLNLPFRDVDWTE
Mflv_3502|M.gilvum_PYR-GCK LMCDVDIDGVISTPDAPSIYDIPKVLHREELDAYVVRRLNLAFRDVDWTQ
Mvan_3283|M.vanbaalenii_PYR-1 LMCDVDIDGVISTPDAPSIYDIPKVLHREELDAYVVRRLNLPFRDVDWTQ
MSMEG_3746|M.smegmatis_MC2_155 LMCDVDVDGVISTPDAPSIYDIPKVLHREELDAYVVRRLNLPFRDVDWTE
TH_1544|M.thermoresistible__bu LMCDVDVDGVISTPDAPSIYDIPKVLHREELDAFVVRRLNLPFRDVDWTQ
******:**:**** ******************:*******.*******:
Mb1725|M.bovis_AF2122/97 WDDLLRRVHEPHETVRIALVGKYVELSDAYLSVAEALRAGGFKHRAKVEI
Rv1699|M.tuberculosis_H37Rv WDDLLRRVHEPHETVRIALVGKYVELSDAYLSVAEALRAGGFKHRAKVEI
MMAR_2504|M.marinum_M WDDLLRRVHEPKETVRIALVGKYVELSDAYLSVIEAIRAGGFKHRAKVEI
MUL_1688|M.ulcerans_Agy99 WDDLLRRVHEPKETVRIALVGKYVELSDAYLSVIEAIRAGGFKHRAKVEI
MLBr_01363|M.leprae_Br4923 WDDLLRRVHEPHGTVRIALVGKYVDFSDAYLSVSEALHAGGFKHYAKVEV
MAV_3072|M.avium_104 WDDLLRRVHEPHETVRIALVGKYVELSDAYLSVTEALRAGGFFHHAKVEM
MAB_2364|M.abscessus_ATCC_1997 WGDLLQRVHEPSESVRIALVGKYIDLPDAYLSVTEALRAGGFRHHTKVDI
Mflv_3502|M.gilvum_PYR-GCK WNDLLHRVHEPRETVRIALVGKYIDLSDAYLSVAEALRAGGFAHHAKVEI
Mvan_3283|M.vanbaalenii_PYR-1 WNDLLRRVHEPRDTVRIALVGKYIDLSDAYLSVAEALRAGGFAQHAKVEM
MSMEG_3746|M.smegmatis_MC2_155 WDDLLRRVHEPQETVRIALVGKYIDLSDAYLSVAEALRAGGFKHRAKVEM
TH_1544|M.thermoresistible__bu WNDLLSRVHDPRETVRIALVGKYIDLSDAYLSVAEALRAGGFAHHARVEM
*.*** ***:* :*********:::.****** **::**** : ::*::
Mb1725|M.bovis_AF2122/97 CWVASDGCETTSGAAAALGDVHGVLIPGGFGIRGIEGKIGAIAYARARGL
Rv1699|M.tuberculosis_H37Rv CWVASDGCETTSGAAAALGDVHGVLIPGGFGIRGIEGKIGAIAYARARGL
MMAR_2504|M.marinum_M SWVGSDDCQTDGGVASALGDVHGVLIPGGFGIRGIEGKISAISYARSRGL
MUL_1688|M.ulcerans_Agy99 SWVGSDDCQTDGGVASALGDVHGVLIPGGFGIRGIEGKISAISYARSRGL
MLBr_01363|M.leprae_Br4923 VWVASDDCETATGAAAVLADVHGVLIPGGFGIRGIEGKIGAIRYARARGL
MAV_3072|M.avium_104 VWVASDDCESASGAAAALGEVHGVLIPGGFGIRGIEGKIGAIHYARTRGL
MAB_2364|M.abscessus_ATCC_1997 KWVPSDDCETESGAQAALGDVDGVLIPGGFGIRGIEGKLGAIRYARKRRV
Mflv_3502|M.gilvum_PYR-GCK RWIPSDDCETDVGAATALSDVDGVLIPGGFGIRGIEGKVGAIRYARKRGL
Mvan_3283|M.vanbaalenii_PYR-1 RWVASDDCETDASAAAALADVDGVLIPGGFGIRGIEGKIGAIHYARKRGL
MSMEG_3746|M.smegmatis_MC2_155 RWVASDDCETEHGAAAALSDVHGVLIPGGFGIRGIEGKIGAISYARKRGL
TH_1544|M.thermoresistible__bu RWVASDDCDSDSGVKAALEDVHGVLIPGGFGIRGIEGKIGAVRYARKRGL
*: **.*:: .. :.* :*.****************:.*: *** * :
Mb1725|M.bovis_AF2122/97 PVLGLCLGLQCIVIEAARSVGLTNANSAEFDPDTPDPVIATMPDQEEIVA
Rv1699|M.tuberculosis_H37Rv PVLGLCLGLQCIVIEAARSVGLTNANSAEFDPDTPDPVIATMPDQEEIVA
MMAR_2504|M.marinum_M PVFGLCLGLQCIVIEAARSVGLTEANSAEFEPGTPDPVISTMADQEHIVS
MUL_1688|M.ulcerans_Agy99 PVFGLCLGLQCIVIEAARSVGLTEANSAEFEPGTPDPVISTMADQEHIVS
MLBr_01363|M.leprae_Br4923 PVLGLCLGLQCIVIEATRSVGLVQANSAEFEPATPDPVISTMADQKEIVA
MAV_3072|M.avium_104 PVLGLCLGLQCIVIEAARAAGLAGANSAEFDPDTPDPVISTMADQVDIVA
MAB_2364|M.abscessus_ATCC_1997 PILGLCLGLQCMVIEAARSAGLADANSAEFDPDTPSPVIATMADQVRAVA
Mflv_3502|M.gilvum_PYR-GCK PLLGLCLGLQCIVIEAARSVGLTEASSAEFDPDTPDPVISTMADQRDAVA
Mvan_3283|M.vanbaalenii_PYR-1 PVLGLCLGLQCIVIEAARSVGLTEANSAEFDPATPDPVISTMADQRDAVA
MSMEG_3746|M.smegmatis_MC2_155 PVLGLCLGLQCIVIEAARSVGITEANSAEFDPKTPDPVISTMADQRDAVA
TH_1544|M.thermoresistible__bu PVLGLCLGLQCIVIEAARSVGLTQANSTEFDPDTPEPVISTMADQLDAVT
*::********:****:*:.*:. *.*:**:* **.***:**.** *:
Mb1725|M.bovis_AF2122/97 GEADLGGTMRLGSYPAVLEPDSVVAQAYQTTQVSERHRHRYEVNNAYRDK
Rv1699|M.tuberculosis_H37Rv GEADLGGTMRLGSYPAVLEPDSVVAQAYQTTQVSERHRHRYEVNNAYRDK
MMAR_2504|M.marinum_M GQADLGGTMRLGAYPAVLESGSIVAEAYQSTKVSERHRHRYEVNNAYRDR
MUL_1688|M.ulcerans_Agy99 GQADLGGTMRLGAYPAVLESGSIVAEAYQSTKVSERHRHRYEVNNAYRER
MLBr_01363|M.leprae_Br4923 GEADFGGTMRLGAYPAVLQPASIVAQAYGTTQVSERHRHRYEVNNAYRDW
MAV_3072|M.avium_104 GAADLGGTMRLGAYPAVLEEDSIVARAYGTTQVSERHRHRYEVNNAYRDK
MAB_2364|M.abscessus_ATCC_1997 GEADLGGTMRLGAYPAVLEPDSIVAEAYGTTEVSERHRHRYEVNNPYRDK
Mflv_3502|M.gilvum_PYR-GCK GEADLGGTMRLGAYPAVLQKNSLVAKAYDATEVSERHRHRYEVNNAYRER
Mvan_3283|M.vanbaalenii_PYR-1 GEADLGGTMRLGAYPAVLEPNSLVAQAYGTIEVSERHRHRYEVNNTYRDR
MSMEG_3746|M.smegmatis_MC2_155 GEADLGGTMRLGAYPAVLTPNSVVAQAYQSTEVSERHRHRFEVNNAYRDR
TH_1544|M.thermoresistible__bu GAADLGGTMRLGAYPAVLEPGSIVAEAYQATEVSERHRHRYEVNNAYRER
* **:*******:***** *:**.** : :********:****.**:
Mb1725|M.bovis_AF2122/97 IAESGLRFSGTSPDGHLVEFVEYPPDRHPFVVGTQAHPELKSRPTRPHPL
Rv1699|M.tuberculosis_H37Rv IAESGLRFSGTSPDGHLVEFVEYPPDRHPFVVGTQAHPELKSRPTRPHPL
MMAR_2504|M.marinum_M IAESGLRFSGTSPDGHLVEFVEYPPEQHPFVVGTQAHPELKSRPTRPHPL
MUL_1688|M.ulcerans_Agy99 IAESGLRFSGTSPDGHLVEFVEYPPEQHPFVVGTQAHPELKSRPTRPHPL
MLBr_01363|M.leprae_Br4923 IAESGLRISGTSPDGYLVEFVEYPANMHPFVVGTQAHPELKSRPTRPHPL
MAV_3072|M.avium_104 IAESGLRFSGTSPDGHLVEFVEYPPDVHPFIVGTQAHPELKSRPTRPHPL
MAB_2364|M.abscessus_ATCC_1997 IAESGLRFSGTSPDGHLVEFVEYPREVHPFAVATQAHPELKSRPTRPHPL
Mflv_3502|M.gilvum_PYR-GCK IAESGLQFSGTSPDGQLVEFVEYPADVHPFLVGTQAHPELKSRPTRPHPL
Mvan_3283|M.vanbaalenii_PYR-1 IAESGLRFSGTSPDGHLVEFVEYPPEVHPFLVGTQAHPELKSRPTRPHPL
MSMEG_3746|M.smegmatis_MC2_155 IAKSGLRFSGTSPDGHLVEFVEYDPQIHPFLVGTQAHPELKSRPTRPHPL
TH_1544|M.thermoresistible__bu IAKSGLRFSGTSPDGHLVEFVEYPREIHPFLVGTQAHPELKSRPTRPHPL
**:***::******* ******* : *** *.*****************
Mb1725|M.bovis_AF2122/97 FVAFVGAAIDYKAGELLP-VEI---PEIPEHTPNGS-SHRDGVGQPLP--
Rv1699|M.tuberculosis_H37Rv FVAFVGAAIDYKAGELLP-VEI---PEIPEHTPNGS-SHRDGVGQPLP--
MMAR_2504|M.marinum_M FAAFVKAAIDYKEGELLP-VEM---PER---VSNGA-ERRDQVGQSIP--
MUL_1688|M.ulcerans_Agy99 FAAFVKAAIDYKEGELLP-VEM---PER---VSNGA-ERRDQVGQSIP--
MLBr_01363|M.leprae_Br4923 FVAFVGAAIDYKSAELLP-VEIPAVPEISEHLPNSSNQHRDGVERSFP--
MAV_3072|M.avium_104 FVAFVGAAMDYKAGELLP-VEI---PEQ----SSNGIQHRDSAARPIP--
MAB_2364|M.abscessus_ATCC_1997 FSAFIRAAMEYKAAERLP-LDP------------------DSE-------
Mflv_3502|M.gilvum_PYR-GCK FVAFVGAAVEYNNGERLP-VDIPAIPT----ADHQSNGAEHALEDAPA--
Mvan_3283|M.vanbaalenii_PYR-1 FAAFVGASLDYKNAERLP-VEMPEIP------EHEPNGAEHALEDAPA--
MSMEG_3746|M.smegmatis_MC2_155 FAAFIGAAIDYKAAERLPGMDLPEQFVPVEHSDADAPALEEPLEKSDV--
TH_1544|M.thermoresistible__bu FTAFVGAALEFKAAERLP-VEIPEHHRANG-AEHEPDAAGPEVADAGVGP
* **: *::::: .* ** ::
Mb1725|M.bovis_AF2122/97 ---EPASRG
Rv1699|M.tuberculosis_H37Rv ---EPASRG
MMAR_2504|M.marinum_M ---EPANRG
MUL_1688|M.ulcerans_Agy99 ---EPANRG
MLBr_01363|M.leprae_Br4923 ---APAARG
MAV_3072|M.avium_104 ---EPAARG
MAB_2364|M.abscessus_ATCC_1997 ---------
Mflv_3502|M.gilvum_PYR-GCK -------RG
Mvan_3283|M.vanbaalenii_PYR-1 -------RG
MSMEG_3746|M.smegmatis_MC2_155 -------RG
TH_1544|M.thermoresistible__bu LLQEPVHRG