For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSGNGPFGFDPEDFDRVAREAGEGLRDALDQISRMFTTSGERAGLAGLLDEFSRFSRPRTEPETTGEKGD GVWAIYTVDDEGGAHIEQVFPTELDALRANASNTDPSRRVRFLPYGIAVSVLDTTPRSPGSATDADSGAA TDD
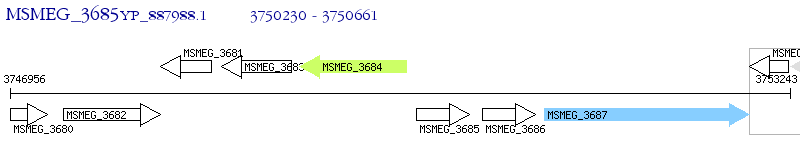
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3685 | - | - | 100% (143) | hypothetical protein MSMEG_3685 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0321 | - | 2e-42 | 66.39% (119) | hypothetical protein Mb0321 |
| M. gilvum PYR-GCK | Mflv_3432 | - | 3e-52 | 72.06% (136) | hypothetical protein Mflv_3432 |
| M. tuberculosis H37Rv | Rv0313 | - | 2e-42 | 66.39% (119) | hypothetical protein Rv0313 |
| M. leprae Br4923 | MLBr_02518 | - | 2e-36 | 58.20% (122) | hypothetical protein MLBr_02518 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0564 | - | 4e-39 | 60.32% (126) | hypothetical protein MMAR_0564 |
| M. avium 104 | MAV_4847 | - | 6e-42 | 61.90% (126) | hypothetical protein MAV_4847 |
| M. thermoresistible (build 8) | TH_0701 | - | 1e-49 | 67.13% (143) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4741 | - | 3e-39 | 60.32% (126) | hypothetical protein MUL_4741 |
| M. vanbaalenii PYR-1 | Mvan_3186 | - | 4e-47 | 67.88% (137) | hypothetical protein Mvan_3186 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0564|M.marinum_M -------------MG-DNGGFGFDPDDFDRVIREGSEGLRDAFERIGRFV
MUL_4741|M.ulcerans_Agy99 -------------MG-DNGGFGFDPDDFDRVIREGSEGLRDAFERIGRFV
Mb0321|M.bovis_AF2122/97 -------------MG-DYGPFGFDPDEFDRVIREGSEGLRDAFERIGRFL
Rv0313|M.tuberculosis_H37Rv -------------MG-DYGPFGFDPDEFDRVIREGSEGLRDAFERIGRFL
MLBr_02518|M.leprae_Br4923 -------------MG-EYSAFGFDPDDFDRLIKEGSEGLRDAFERISRFV
MAV_4847|M.avium_104 -------------MG-EQGGFGFDPDEFDRMIREGSEGLREVFERVSKFV
Mflv_3432|M.gilvum_PYR-GCK -------------MSNSNGPFGFDPEDLDRMVREAGEGLRDALEGLGRFV
Mvan_3186|M.vanbaalenii_PYR-1 -------------MSSSNGPFGFDPEDFERVVREAGEGLREALG------
MSMEG_3685|M.smegmatis_MC2_155 -------------MS-GNGPFGFDPEDFDRVAREAGEGLRDALDQISRMF
TH_0701|M.thermoresistible__bu VCSGSRQRHKEDAMA-NNGPFGFDPDDFDRVAREA-------LDGISRLF
*. . *****::::*: :*. :
MMAR_0564|M.marinum_M SGPGDRPAWSMIFEDLGHRRRP--APETAGEAGDGVWAIYTVDADGGARV
MUL_4741|M.ulcerans_Agy99 SGPGDRPAWSMIFEDLGHRRRP--APETAGEAGDGVWAIYTVDADGGARV
Mb0321|M.bovis_AF2122/97 SSSGAGTGWSAIFEDLSRRSRP--APETAGEAGDGVWAIYTVDADGGARV
Rv0313|M.tuberculosis_H37Rv SSSGAGTGWSAIFEDLSRRSRP--APETAGEAGDGVWAIYTVDADGGARV
MLBr_02518|M.leprae_Br4923 GGPGVRTAWSAIFEDLSRRARP--AQETADEAGDGVWAIYTVTGDGAARV
MAV_4847|M.avium_104 AAPGARTGWSSLFEDLSRRGRP--APETAGEAGDGVWAIYTVDAGGGAHV
Mflv_3432|M.gilvum_PYR-GCK NTPGERTGWSVLFDEFSRGTRPRTRPETTGEAGDGVWAIFTVDGEGGARI
Mvan_3186|M.vanbaalenii_PYR-1 -----RAGWSTLFDEFSRGARPRTQPETTGEAGDGVWAIYTTDATGGAHI
MSMEG_3685|M.smegmatis_MC2_155 TTSGERAGLAGLLDEFSRFSRPRTEPETTGEKGDGVWAIYTVDDEGGAHI
TH_0701|M.thermoresistible__bu TTAGERAGWANLFDDIARRGRGRTEPETTGETGDGVWVIYTVDADGGAHI
.. : :::::.: * **:.* *****.*:*. *.*::
MMAR_0564|M.marinum_M EQVYATELDALRANKDNTDSRRKVRFLPYGIAVSVLDD------------
MUL_4741|M.ulcerans_Agy99 EQVYATELDALRANKDNTDSRRKVRFLPYGIAVSVLDD------------
Mb0321|M.bovis_AF2122/97 EQVYATELDALRANKDNTDPKRKVRFLPYGIAVSVLDD------------
Rv0313|M.tuberculosis_H37Rv EQVYATELDALRANKDNTDPKRKVRFLPYGIAVSVLDD------------
MLBr_02518|M.leprae_Br4923 EQVYATELDALRANKNNVDPKRKVRFLPYGIAVSVLDS------------
MAV_4847|M.avium_104 EQVFATELDALRANKDNVDPKRKVRFLPYGIAVSVLDD------------
Mflv_3432|M.gilvum_PYR-GCK EQVYPTELDALRANKDNTDPTRRVRFLPYGIAVSVLDS------------
Mvan_3186|M.vanbaalenii_PYR-1 EQVYPTELDALRANKDNVDPKRRVRFLPYGIAVSVLDAAAGGGESESAAG
MSMEG_3685|M.smegmatis_MC2_155 EQVFPTELDALRANASNTDPSRRVRFLPYGIAVSVLDTT-----------
TH_0701|M.thermoresistible__bu EQVFPTELDALRAHRDNTDPARKVRFLPYGIAASVLDAP-----------
***:.********: .*.*. *:*********.****
MMAR_0564|M.marinum_M ----------SSGEPE---
MUL_4741|M.ulcerans_Agy99 ----------SSGEPE---
Mb0321|M.bovis_AF2122/97 ----------PVDEAQ---
Rv0313|M.tuberculosis_H37Rv ----------PVDEAQ---
MLBr_02518|M.leprae_Br4923 ----------HQESTQQL-
MAV_4847|M.avium_104 ----------DPEQPG---
Mflv_3432|M.gilvum_PYR-GCK GPDSNPDAG-VAGSPDQHS
Mvan_3186|M.vanbaalenii_PYR-1 GGETNPADGDDSGSADQHP
MSMEG_3685|M.smegmatis_MC2_155 PRSPGSATDADSGAATDD-
TH_0701|M.thermoresistible__bu SPREQSGSPDEPGDTADDR
.