For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGDDHGHEHGPRRELPPGLAEQLDLPYAGMVSFGHRPFLTETQQLDEWRPDVAIVGAPFDIATTNRPGAR FGPRAIRATAYEPGTYHLDLGLEIFDWLEVVDFGDAYCPHGQTEVSHANIRERVHAVASRGVVPVVLGGD HSITWPSATAVADVHGYGNVGIVHFDAHADTADNIEGNLASHGTPMRRLIESGAVPGSHFVQVGLRGYWP PQDTFEWMLEQKMTWHTMQEIWDRGFKAVMDDAVGEALAKAEKLYLSVDIDVLDPAHAPGTGTPEPGGIT SADLLRMVRQLCHRHDVAGVDVVEVAPAYDHAELTVNAAHRVVFEALAGMAARRRDAAQAQPGPPAR
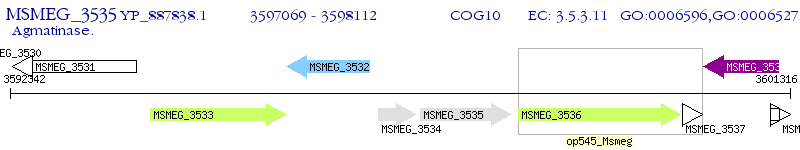
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3535 | speB | - | 100% (347) | agmatinase |
| M. smegmatis MC2 155 | MSMEG_4374 | speB | 9e-53 | 41.25% (320) | agmatinase |
| M. smegmatis MC2 155 | MSMEG_1072 | speB | 2e-47 | 38.59% (311) | agmatinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3303 | - | 0.0 | 90.58% (329) | putative agmatinase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3257 | - | 7e-78 | 49.07% (324) | PUTATIVE putative agmatinase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3024 | - | 0.0 | 90.35% (342) | putative agmatinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3303|M.gilvum_PYR-GCK ---------------------MAEQLELAYAGMASFGHRPFLTEVEQLDS
Mvan_3024|M.vanbaalenii_PYR-1 -----MGHDHRPHRELAPG--MAEQLDLPYAGVVSFGHRPFLTESEQLDS
MSMEG_3535|M.smegmatis_MC2_155 -MGDDHGHEHGPRRELPPG--LAEQLDLPYAGMVSFGHRPFLTETQQLDE
TH_3257|M.thermoresistible__bu VWRQPVDRSRDPHREPGPINLQRHQFTPAYSGIATLFGLPLCLNQDDLRA
.*: .*:*:.:: *: : ::*
Mflv_3303|M.gilvum_PYR-GCK WRPDAAIVGAPFDVGTTNRPGARFGPRAIRATA-YEPGTYHMDLG----L
Mvan_3024|M.vanbaalenii_PYR-1 WKPDVAVVGAPFDVGTTNRPGARFGPRAIRATA-YEPGTYHMDLG----L
MSMEG_3535|M.smegmatis_MC2_155 WRPDVAIVGAPFDIATTNRPGARFGPRAIRATA-YEPGTYHLDLG----L
TH_3257|M.thermoresistible__bu GAVDVAVVGAPVDVSLGHR-GAAYGPRAIRADERVLPHTPTMLINDSTRV
*.*:****.*:. :* ** :******* * * : :. :
Mflv_3303|M.gilvum_PYR-GCK EIFDWLEVVDFGDAYCPHGQTEVSHNNIRERVHMLASRGIVPVVLGGDHS
Mvan_3024|M.vanbaalenii_PYR-1 EIFDWLEVVDFGDAYCPHGQTEVSHRNIRERVHAVADRGIVPVILGGDHS
MSMEG_3535|M.smegmatis_MC2_155 EIFDWLEVVDFGDAYCPHGQTEVSHANIRERVHAVASRGVVPVVLGGDHS
TH_3257|M.thermoresistible__bu KPFEVLNVVDYGDAAVDPLSIENSMGPIRELVRDVAEVGAVPLVLGGDHS
: *: *:***:*** . * * *** *: :*. * **::******
Mflv_3303|M.gilvum_PYR-GCK ITWPAATAVADVHGYGNVGIVHFDAHADTADEIEGNLASHGTPMRRLIES
Mvan_3024|M.vanbaalenii_PYR-1 ITWPAATAVADVHGYGNVGIVHFDAHADTADEIEGNLASHGTPMRRLIES
MSMEG_3535|M.smegmatis_MC2_155 ITWPSATAVADVHGYGNVGIVHFDAHADTADNIEGNLASHGTPMRRLIES
TH_3257|M.thermoresistible__bu ILWPNAAAIADVYGAGKVGVVHFDAHPDCADDVVGHHASHGTPIRRLIDD
* ** *:*:***:* *:**:******.* **:: *: ******:****:.
Mflv_3303|M.gilvum_PYR-GCK GAVPGSHFVQVGLRGYWPP-RDTFDWMLEQKMTWHTMQEIWERGFKAVMA
Mvan_3024|M.vanbaalenii_PYR-1 GAVPGSHFVQVGLRGYWPP-QDTFEWMLEQKMTWHTMQEIWERGFKAVMA
MSMEG_3535|M.smegmatis_MC2_155 GAVPGSHFVQVGLRGYWPP-QDTFEWMLEQKMTWHTMQEIWDRGFKAVMD
TH_3257|M.thermoresistible__bu EHIPGRNFIQVGLRSAIAPDEELFAWMRDNGLRTHFMAEIDRRGFHTVLQ
:** :*:*****. .* .: * ** :: : * * ** ***::*:
Mflv_3303|M.gilvum_PYR-GCK DAVGEALAKADKLYVSVDIDVLDPAHAPGTGTPEPGGITSADLLRMVRQL
Mvan_3024|M.vanbaalenii_PYR-1 DAVAEALAKADKLYVSVDIDVLDPAHAPGTGTPEPGGITSADLLRMVRQL
MSMEG_3535|M.smegmatis_MC2_155 DAVGEALAKAEKLYLSVDIDVLDPAHAPGTGTPEPGGITSADLLRMVRQL
TH_3257|M.thermoresistible__bu DAIDEALDGPEYLYLSIDIDVLDPAYAPGTGTPEPPGLTNRELLPAIRRI
**: *** .: **:*:********:********* *:*. :** :*::
Mflv_3303|M.gilvum_PYR-GCK CYEHDVAGVDVVEVAPAYDHAELTVNAAHRVVFEALAGMAARCRDAANGE
Mvan_3024|M.vanbaalenii_PYR-1 CHEHDVVGVDVVEVAPAYDHAELTINAAHRVVFEALAGMAARRRDAADGE
MSMEG_3535|M.smegmatis_MC2_155 CHRHDVAGVDVVEVAPAYDHAELTVNAAHRVVFEALAGMAARRRDAAQAQ
TH_3257|M.thermoresistible__bu CHETPVVGLEIVEVAPHLDPGYTTTMNARRVIFEALTGLAMRRLGLPGPD
*:. *.*:::***** * . * *:**:****:*:* * . . :
Mflv_3303|M.gilvum_PYR-GCK VGQPARSYRDRDATSRE
Mvan_3024|M.vanbaalenii_PYR-1 VGQPARSYRDRGVTSPE
MSMEG_3535|M.smegmatis_MC2_155 PGPPAR-----------
TH_3257|M.thermoresistible__bu YLHPDVAGPPSNRV---
*