For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSPHRDGMTATTDAPSTLTPARISRLRVPELEELQNANVRGFFTRQQREEGLTSNWFRALSLNEDDLGRL NGYLLPILGADGRDGLTAREREVIATVVSGENRCAYCHTNHANKLGKVAGDWSFGQRVAIDHTQVGELTD RERALGDLAVLVNNHPQQIRDEDFDRLRRVGLDDHLILEAISIAAFIGATNRIGIALSVPPNPEYTGIPQ
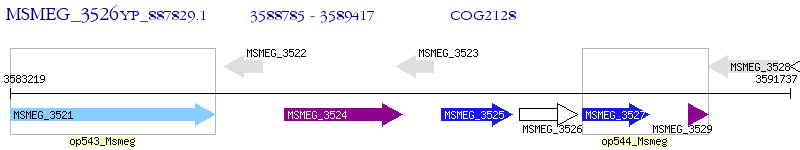
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3526 | - | - | 100% (210) | hypothetical protein MSMEG_3526 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3311c | - | 7e-06 | 28.57% (175) | hypothetical protein MAB_3311c |
| M. marinum M | MMAR_5256 | - | 9e-06 | 28.69% (122) | hypothetical protein MMAR_5256 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4333 | - | 8e-06 | 28.69% (122) | hypothetical protein MUL_4333 |
| M. vanbaalenii PYR-1 | Mvan_3017 | - | 3e-90 | 81.68% (202) | uncharacterized peroxidase-related enzyme |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3526|M.smegmatis_MC2_155 MSPHRDGMTATTDAPSTLTPARISRLRVPELEELQNANVRGFFTRQQREE
Mvan_3017|M.vanbaalenii_PYR-1 -------MTATADAP-VQAPATISRLAVPELDELRSPGVRGFFNRQLRDE
MMAR_5256|M.marinum_M --------------------MRLPSITRGRPGRRPVFALARFVSGRDIPD
MUL_4333|M.ulcerans_Agy99 --------------------MRLPSITRGRPGRRPVFALARFVSGREIPD
MAB_3311c|M.abscessus_ATCC_199 ----------------------MSRLTTPASEDITAEAESALGTVG-RQF
:. : : .
MSMEG_3526|M.smegmatis_MC2_155 GLTSNWFRALSLNEDDLGRLNGYLLPILGADGRDGLTAREREVIATVVSG
Mvan_3017|M.vanbaalenii_PYR-1 GLTSNWFRALSLNEGDLERLNAYLLPLLGTDGRGGLSAREREVIATVVSG
MMAR_5256|M.marinum_M IIKVRYYRPRFFGKPFFQLAQALLR------GPSPWTIGERELFAAFVSA
MUL_4333|M.ulcerans_Agy99 IIKVRYYRSRFFGKPFFQLAQALLR------GPSPWTIGERELFAAFVSA
MAB_3311c|M.abscessus_ATCC_199 GFVPNMFAMLASNPAVLDVVMSLQG-----SLGRVLDAKTRHTIALAVSE
: . : . : . *. :* **
MSMEG_3526|M.smegmatis_MC2_155 ENRCAYCHTNHANKLGKVAGDWSFGQRVAIDHTQVGELTDRERALGD-LA
Mvan_3017|M.vanbaalenii_PYR-1 ENRCAYCHTNHANKLGKVAGDWSFGQRVAIDHHQVAELTERERALAD-LA
MMAR_5256|M.marinum_M RNRCQFCATTHEAVAAATLDPD--LAAAVVSDWRTAPVDEPVRATLG-LL
MUL_4333|M.ulcerans_Agy99 RNRCQFCATTHEAVAAATLDPG--LAAAVVSDWRTAPVDEPLRATLG-LL
MAB_3311c|M.abscessus_ATCC_199 ANGCDYCLAVHSYVSAELGGMS----SDDIDLARSGSSIDPKRAAVARFA
* * :* : * . . :. : . : ** :
MSMEG_3526|M.smegmatis_MC2_155 VLVNNHPQQIRDEDFDRLRRVGLDDHLILEAISIAAFIGATNRIG--IAL
Mvan_3017|M.vanbaalenii_PYR-1 VAVNSDPQSIRDSDIVRLRDLGLDDHEILEGISIAAFIGATNRIG--IAL
MMAR_5256|M.marinum_M DKLTLTPAQVRPVDVRAVLDAGVSAEAIRDAIEICALFNTINRVADGLDF
MUL_4333|M.ulcerans_Agy99 DKLTLTPAQVRPVDVRAVLDAGVSAEAIRDAIEICALFNTINRVADGLDF
MAB_3311c|M.abscessus_ATCC_199 QRVVETRGKVGDSDLAAVRGAGYADSQILAIVTVAVQTLLTNYINNVNQT
: .: *. : * * : :.. * :
MSMEG_3526|M.smegmatis_MC2_155 SVPPNPEYTGIPQ--------
Mvan_3017|M.vanbaalenii_PYR-1 SVPPNPEYTGIPAGRGHTTA-
MMAR_5256|M.marinum_M ALPTGTERTLNTRVLLRLGYR
MUL_4333|M.ulcerans_Agy99 ALPTGTERTLNTRVLLRLGYR
MAB_3311c|M.abscessus_ATCC_199 VIDIPAAGTAA----------
: . *