For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ANAEPTTPVPAPAPAPSPAVGTGTGPAPGPAPEAAAAEGPAPVMDHDGIYVVGTDIQPGTYATGGPADGQ TCYWKRMADLHGGDIIDNAFTKKPDVVQIEATDKAFKTSGCQPWTQTTAAPTDTPTGEGAPGDLPGLLAK AKLRTWLDTINNGARQYDGSQVPLP*
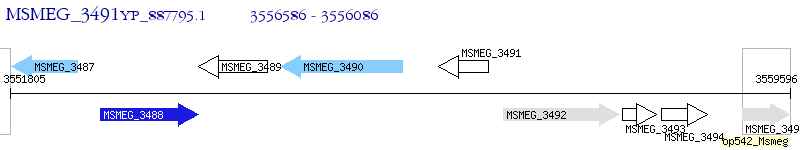
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3491 | - | - | 100% (166) | hypothetical protein MSMEG_3491 |
| M. smegmatis MC2 155 | MSMEG_1588 | - | 1e-10 | 34.91% (106) | hypothetical protein MSMEG_1588 |
| M. smegmatis MC2 155 | MSMEG_5153 | - | 3e-07 | 40.45% (89) | hypothetical protein MSMEG_5153 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1941c | - | 5e-29 | 44.71% (170) | hypothetical protein Mb1941c |
| M. gilvum PYR-GCK | Mflv_1068 | - | 2e-24 | 49.14% (116) | hypothetical protein Mflv_1068 |
| M. tuberculosis H37Rv | Rv1906c | - | 5e-29 | 44.71% (170) | hypothetical protein Rv1906c |
| M. leprae Br4923 | MLBr_02010 | - | 2e-30 | 47.10% (155) | putative lipoprotein |
| M. abscessus ATCC 19977 | MAB_1945c | - | 8e-06 | 48.84% (43) | dihydrolipoamide succinyltransferase |
| M. marinum M | MMAR_2802 | - | 2e-31 | 53.68% (136) | hypothetical protein MMAR_2802 |
| M. avium 104 | MAV_2795 | - | 2e-35 | 51.97% (152) | hypothetical protein MAV_2795 |
| M. thermoresistible (build 8) | TH_0862 | - | 6e-37 | 49.42% (172) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2927 | - | 9e-31 | 54.07% (135) | hypothetical protein MUL_2927 |
| M. vanbaalenii PYR-1 | Mvan_5756 | - | 1e-25 | 48.82% (127) | hypothetical protein Mvan_5756 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1068|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5756|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3491|M.smegmatis_MC2_155 --------------------------------------------------
TH_0862|M.thermoresistible__bu --------------------------------------------------
MMAR_2802|M.marinum_M --------------------------------------------------
MUL_2927|M.ulcerans_Agy99 --------------------------------------------------
Mb1941c|M.bovis_AF2122/97 --------------------------------------------------
Rv1906c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02010|M.leprae_Br4923 --------------------------------------------------
MAV_2795|M.avium_104 --------------------------------------------------
MAB_1945c|M.abscessus_ATCC_199 MAFSVQMPALGESVTEGTVTRWLKQEGDTVEVDEPLLEVSTDKVDTEIPA
Mflv_1068|M.gilvum_PYR-GCK -------------------MRLTRPAALGAAAAASCLLLAPVASAQPEPT
Mvan_5756|M.vanbaalenii_PYR-1 -------------------MRLRKPAVLVTAALAGWCVLAPVAGAQPAPE
MSMEG_3491|M.smegmatis_MC2_155 ----------------------MANAEPTTPVPAPAPAPSPAVGTGTGPA
TH_0862|M.thermoresistible__bu --------------MRSRTAALSAVAALVAGVWLTSPWPAGAEPTIEPAE
MMAR_2802|M.marinum_M --------MPAVRNALLTPVGCSAMGSKVMASHAARLGAAGLLLAGWAGS
MUL_2927|M.ulcerans_Agy99 ------------------------MGSKVMASHAARLGAAGLLLAGWA-A
Mb1941c|M.bovis_AF2122/97 ------------------------MRLKPAPSPAAAFAVAGLILAGWAGS
Rv1906c|M.tuberculosis_H37Rv ------------------------MRLKPAPSPAAAFAVAGLILAGWAGS
MLBr_02010|M.leprae_Br4923 ------------------------MWLTAAP-AAARFVVASVIAASCAAS
MAV_2795|M.avium_104 -------MAAGWAASTVVAGADPDETHGPAPGPAATPSPAPSASPSPAAS
MAB_1945c|M.abscessus_ATCC_199 PTSGVLTKIVAREDDTVEIGGELGVISEAGEAAPAAPAPAPAPAAAPAPE
: .
Mflv_1068|M.gilvum_PYR-GCK P-------APDAEGPKTTID---ASGSYAVGTEILPGVYQSAGPVDDGAC
Mvan_5756|M.vanbaalenii_PYR-1 PPPAPPADAPPAEGPKTTID---ANGSYAVGTEIAPGVYQSAGPIDGGAC
MSMEG_3491|M.smegmatis_MC2_155 PG--PAPEAAAAEGPAPVMD---HDGIYVVGTDIQPGTYATGGPADGQTC
TH_0862|M.thermoresistible__bu PSGEPAPQAAPAQ-PRTTID---ADGTYAVGTDVVPGTYSSAGPVEGDVC
MMAR_2802|M.marinum_M MGVAAADPQPAPPAPKTTMD---HDGTYAVGTDIAPGTYTSAGPVGDGTC
MUL_2927|M.ulcerans_Agy99 MGVAAADPQPAPPALKTTMD---HDGTYAVGTDIVPGTYTSAGPVGDGTC
Mb1941c|M.bovis_AF2122/97 VGLAGADPEPAP-TPKTAID---SDGTYAVGIDIAPGTYSSAGPVGDGTC
Rv1906c|M.tuberculosis_H37Rv VGLAGADPEPAP-TPKTAID---SDGTYAVGIDIAPGTYSSAGPVGDGTC
MLBr_02010|M.leprae_Br4923 TGVAGADPQSPS-APKTTID---HDGTYAVGTDIAPGTYSSAGPVGNGTC
MAV_2795|M.avium_104 PSPS-ASPAPAPAAPKTTID---KDGIYAVGTDIAPGVYSSGGPVGNGTC
MAB_1945c|M.abscessus_ATCC_199 PTPAPAPEATATAAPEPAAEPPAAAAPAGSGTSVKMPELGESVTEGTVTR
. . .. : . * .: . . .
Mflv_1068|M.gilvum_PYR-GCK YWKRVAGEE-------------------------------LVDNALTKKP
Mvan_5756|M.vanbaalenii_PYR-1 YWKRTAGEE-------------------------------TVDNRLTKKP
MSMEG_3491|M.smegmatis_MC2_155 YWKRMADLH----------------------------GGDIIDNAFTKKP
TH_0862|M.thermoresistible__bu YWKRVSSADGQAD------------------------GGKILDNAMSKKP
MMAR_2802|M.marinum_M YWKRMSNPD-----------------------------GALIDNAMTKKP
MUL_2927|M.ulcerans_Agy99 YWKRMSNPD-----------------------------GALIDNAMTKKP
Mb1941c|M.bovis_AF2122/97 YWKRMGNPD-----------------------------GALIDNALSKKP
Rv1906c|M.tuberculosis_H37Rv YWKRMGNPD-----------------------------GALIDNALSKKP
MLBr_02010|M.leprae_Br4923 YWKRIDNPD-----------------------------GP-IDNAMSKKP
MAV_2795|M.avium_104 YWKRMGNPD-----------------------------GAVIDNAMTKKP
MAB_1945c|M.abscessus_ATCC_199 WLKKVGDEVGVDEPLVEVSTDKVDTEIPSPVAGVLLSISANEDDTVAVGG
: *: . *: .:
Mflv_1068|M.gilvum_PYR-GCK AFVQIMPTDTTFTTSDCQAWTLTNAPMPP----------QPSPAS-----
Mvan_5756|M.vanbaalenii_PYR-1 SFVQIMPTDTTFTTSDCQTWQLTNAPMPP----------EPGPGS-----
MSMEG_3491|M.smegmatis_MC2_155 DVVQIEATDKAFKTSGCQPWTQTTAAPTDTPTGEGAPGDLPGLLAKAKLR
TH_0862|M.thermoresistible__bu QVVQIEATDGAFVTRGCQPWRQTTAPSGE----IDLPPEVDALIGKAKLR
MMAR_2802|M.marinum_M QVVQIEPTDKAFKTSGCQPWQST-PDADV-------PAELPGPAAGGQLQ
MUL_2927|M.ulcerans_Agy99 QVVQIEPTDKAFKTSGCQPWQST-PDADV-------PAELPGPAAGGQLQ
Mb1941c|M.bovis_AF2122/97 QVVTIEPTDKAFKTHGCQPWQNTGSEGAA-------PAGVPGPEAGAQLQ
Rv1906c|M.tuberculosis_H37Rv QVVTIEPTDKAFKTHGCQPWQNTGSEGAA-------PAGVPGPEAGAQLQ
MLBr_02010|M.leprae_Br4923 KIVQIEASNKAFKTTGCQPWQQT-SNTTV-------STDLPGPIAGIQLE
MAV_2795|M.avium_104 QVVQIDPGDKSFKTSGCQPWQLT-PDAVP-------PPSAP-PAG---VQ
MAB_1945c|M.abscessus_ATCC_199 ELAVVGAADAAPAAPAAPAAPAPEPAAAPAAAAAPPAPPAPAPAPAAPAP
.. : . : : : . . . .
Mflv_1068|M.gilvum_PYR-GCK -LLGELTKAIGSGMFSGGPPP-----------------------------
Mvan_5756|M.vanbaalenii_PYR-1 -LLGELTQAIGSGMFSGGPR------------------------------
MSMEG_3491|M.smegmatis_MC2_155 TWLDTINNGARQYDGSQVPLP-----------------------------
TH_0862|M.thermoresistible__bu HYIDTLNRNAQAFG--QVPPP-----------------------------
MMAR_2802|M.marinum_M MNLGILN-GLLAPNGQQVPPS-----------------------------
MUL_2927|M.ulcerans_Agy99 MNLGILN-GLLAPNGQQVPPS-----------------------------
Mb1941c|M.bovis_AF2122/97 NQLGILN-GLLGPTGGRVPQP-----------------------------
Rv1906c|M.tuberculosis_H37Rv NQLGILN-GLLGPTGGRVPQP-----------------------------
MLBr_02010|M.leprae_Br4923 SNLGILN-GLLASNGQQVPRS-----------------------------
MAV_2795|M.avium_104 GTLGILN-GLLGGAGQQAPSAPSAPAAPPSGAQSPAAAPAPQAAHP----
MAB_1945c|M.abscessus_ATCC_199 ASTAPAANGAAGDNPYVTPLVRKLAAENNVDLSALSGSGVGGRIRKQDVL
*
Mflv_1068|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5756|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3491|M.smegmatis_MC2_155 --------------------------------------------------
TH_0862|M.thermoresistible__bu --------------------------------------------------
MMAR_2802|M.marinum_M --------------------------------------------------
MUL_2927|M.ulcerans_Agy99 --------------------------------------------------
Mb1941c|M.bovis_AF2122/97 --------------------------------------------------
Rv1906c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02010|M.leprae_Br4923 --------------------------------------------------
MAV_2795|M.avium_104 --------------------------------------------------
MAB_1945c|M.abscessus_ATCC_199 AAAEAAKAPAAAPAPAAPAASAPAAVAPSLAHLRGTTQKANRIRQITAKK
Mflv_1068|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5756|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3491|M.smegmatis_MC2_155 --------------------------------------------------
TH_0862|M.thermoresistible__bu --------------------------------------------------
MMAR_2802|M.marinum_M --------------------------------------------------
MUL_2927|M.ulcerans_Agy99 --------------------------------------------------
Mb1941c|M.bovis_AF2122/97 --------------------------------------------------
Rv1906c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02010|M.leprae_Br4923 --------------------------------------------------
MAV_2795|M.avium_104 --------------------------------------------------
MAB_1945c|M.abscessus_ATCC_199 TRESLQETAQLTQTHEVDVTKIAALRARAKAAFAEREGVNLTFLPFFAKA
Mflv_1068|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5756|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3491|M.smegmatis_MC2_155 --------------------------------------------------
TH_0862|M.thermoresistible__bu --------------------------------------------------
MMAR_2802|M.marinum_M --------------------------------------------------
MUL_2927|M.ulcerans_Agy99 --------------------------------------------------
Mb1941c|M.bovis_AF2122/97 --------------------------------------------------
Rv1906c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02010|M.leprae_Br4923 --------------------------------------------------
MAV_2795|M.avium_104 --------------------------------------------------
MAB_1945c|M.abscessus_ATCC_199 AVEALKTHPNVNASYNEESKEITYFDAEHLGIAVDTDQGLLSPVIHNAGD
Mflv_1068|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5756|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3491|M.smegmatis_MC2_155 --------------------------------------------------
TH_0862|M.thermoresistible__bu --------------------------------------------------
MMAR_2802|M.marinum_M --------------------------------------------------
MUL_2927|M.ulcerans_Agy99 --------------------------------------------------
Mb1941c|M.bovis_AF2122/97 --------------------------------------------------
Rv1906c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02010|M.leprae_Br4923 --------------------------------------------------
MAV_2795|M.avium_104 --------------------------------------------------
MAB_1945c|M.abscessus_ATCC_199 LSLAGLARAIADIAARARSGNLKPDELAGGTFTITNIGSQGALFDTPILV
Mflv_1068|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5756|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_3491|M.smegmatis_MC2_155 --------------------------------------------------
TH_0862|M.thermoresistible__bu --------------------------------------------------
MMAR_2802|M.marinum_M --------------------------------------------------
MUL_2927|M.ulcerans_Agy99 --------------------------------------------------
Mb1941c|M.bovis_AF2122/97 --------------------------------------------------
Rv1906c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02010|M.leprae_Br4923 --------------------------------------------------
MAV_2795|M.avium_104 --------------------------------------------------
MAB_1945c|M.abscessus_ATCC_199 PPQAAMLGTGAIVKRPRVIRDDAGNESIGIRSVCYLPLTYDHRLIDGADA
Mflv_1068|M.gilvum_PYR-GCK ----------------------
Mvan_5756|M.vanbaalenii_PYR-1 ----------------------
MSMEG_3491|M.smegmatis_MC2_155 ----------------------
TH_0862|M.thermoresistible__bu ----------------------
MMAR_2802|M.marinum_M ----------------------
MUL_2927|M.ulcerans_Agy99 ----------------------
Mb1941c|M.bovis_AF2122/97 ----------------------
Rv1906c|M.tuberculosis_H37Rv ----------------------
MLBr_02010|M.leprae_Br4923 ----------------------
MAV_2795|M.avium_104 ----------------------
MAB_1945c|M.abscessus_ATCC_199 GRFLTTIKHRLEEGAFEADLGL