For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSHADVPAAVTALRRTQTIRERARFLYHRARDGGSAWFTVDDDALERAASEVAAVTRDNYPDLKIPYHSR WRHFEAGGIDRRQQIGARVSGGDALTRAMVDLTVVSVLLDAGAGPDWRYTEPGTGQTFTRSEGLGVASLH AFCDGLFSSDPHDPLRVDAAGLARLDTHALGRAFQVGPGNPLVGLDGRVELLHRLGGALTEVSGAARPSA LLEPLLGRGPVAAHDILDRLLASLSGIWLTPSAIEGHPLGDCWPHDAVPGPGPSAGWMPFHKLSQWLTYS LLEPFEWAGERITGLDELTGLPEYRNGGLFLDTGVLRLRDDNLTERAWSVGDQLIIEWRALTVALLDEIA PLVRELLGVPDLPLACVLEGGTWAAGRALAQRLRGGRPPLVIDSDGTVF
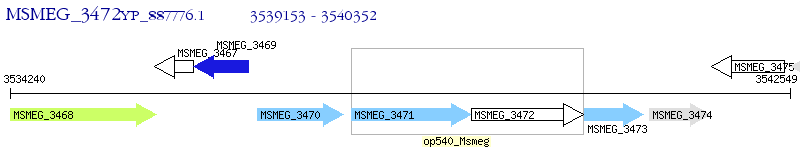
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3472 | - | - | 100% (399) | hypothetical protein MSMEG_3472 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3258 | - | 1e-136 | 62.50% (400) | hypothetical protein Mflv_3258 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2975 | - | 1e-139 | 61.74% (413) | hypothetical protein Mvan_2975 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3258|M.gilvum_PYR-GCK ----------MTSAAEAVAALRNTDAVRTRSAQLLDRARRGRSRWFTVHD
Mvan_2975|M.vanbaalenii_PYR-1 MTVPAHAELDATRPADAVALLRTTDAVRRRSAQLLARARSGESRWFVVDH
MSMEG_3472|M.smegmatis_MC2_155 --------MSHADVPAAVTALRRTQTIRERARFLYHRARDGGSAWFTVDD
: . **: ** *:::* *: * *** * * **.*..
Mflv_3258|M.gilvum_PYR-GCK DALPAAAEDVVAVTRTRYPNLEIPFHSRWRHFDAGGVDRGAELDALLAGT
Mvan_2975|M.vanbaalenii_PYR-1 DAMDGAADDVATVTRERFPDMRIPWHSRWRHFEAGGVDRRAELDSLLGDA
MSMEG_3472|M.smegmatis_MC2_155 DALERAASEVAAVTRDNYPDLKIPYHSRWRHFEAGGIDRRQQIGARVS--
**: **.:*.:*** .:*::.**:*******:***:** ::.: :.
Mflv_3258|M.gilvum_PYR-GCK DDAERARARIDLAVVSVLLDAGAGADWGYMETRGDEVVRYTRSEGLGVAS
Mvan_2975|M.vanbaalenii_PYR-1 GGDERARAMIDLTVVSVLLDAGAGADWRYVETHGDSRRTFARSEGLAVAS
MSMEG_3472|M.smegmatis_MC2_155 GGDALTRAMVDLTVVSVLLDAGAGPDWRYTEPG--TGQTFTRSEGLGVAS
.. :** :**:***********.** * *. ::*****.***
Mflv_3258|M.gilvum_PYR-GCK WHGFVSGVFSGDPHRPLRVDADGLRGLTEERLAEMLQVRPDNPLVGLAGR
Mvan_2975|M.vanbaalenii_PYR-1 WHAFVSGMFSGDAARPLRVDADALSTLDKERLGAALQVSPGNPLVGLDGR
MSMEG_3472|M.smegmatis_MC2_155 LHAFCDGLFSSDPHDPLRVDAAGLARLDTHALGRAFQVGPGNPLVGLDGR
*.* .*:**.*. ****** .* * . *. :** *.****** **
Mflv_3258|M.gilvum_PYR-GCK VTLMHRLGDALASQPGDFGDEGRPGGLFDLLGRGGTVRAHDILTRLLVTL
Mvan_2975|M.vanbaalenii_PYR-1 VALMRSLGDALAAQPEVFGERGRPGFLFDRLAADGTVHAHDILVQLLASL
MSMEG_3472|M.smegmatis_MC2_155 VELLHRLGGALTEVSG----AARPSALLEPLLGRGPVAAHDILDRLLASL
* *:: **.**: . .**. *:: * *.* ***** :**.:*
Mflv_3258|M.gilvum_PYR-GCK SPIWPSDNRIGEVPVGDCWRHDAVQGPGLTAGWVPLHKLSQWLTYSLIEP
Mvan_2975|M.vanbaalenii_PYR-1 SRIWPSRNVIGEDSVGDCWRHDAVTGPGLTAGWLPLHKLSQWLTYSLLEP
MSMEG_3472|M.smegmatis_MC2_155 SGIWLTPSAIEGHPLGDCWPHDAVPGPGPSAGWMPFHKLSQWLTYSLLEP
* ** : . * .:**** **** *** :***:*:***********:**
Mflv_3258|M.gilvum_PYR-GCK FVWAGVEVLDIDALTGLPEYRNGGLLLDTGVLSLRDQHIAARTWNPDDEL
Mvan_2975|M.vanbaalenii_PYR-1 FDWAGVEVTGVGALTGLPEYRNGGLLLDHGVLRLRDPEAATRTWGPGDEL
MSMEG_3472|M.smegmatis_MC2_155 FEWAGERITGLDELTGLPEYRNGGLFLDTGVLRLRDDNLTERAWSVGDQL
* *** .: .:. ************:** *** *** . : *:*. .*:*
Mflv_3258|M.gilvum_PYR-GCK VVEWRALTVALLDELAPLVRAGLG--DPQLPLACVLEGGTWAAGRASAQR
Mvan_2975|M.vanbaalenii_PYR-1 VVEWRALTVALLDELAPLVRDRLGVDAEQMPLACVLEGGTWAAGRGSAQR
MSMEG_3472|M.smegmatis_MC2_155 IIEWRALTVALLDEIAPLVRELLG--VPDLPLACVLEGGTWAAGRALAQR
::************:***** ** ::***************. ***
Mflv_3258|M.gilvum_PYR-GCK LRGGSPPLQVTSDGTVF
Mvan_2975|M.vanbaalenii_PYR-1 LRGGLPPLSVTSDGTVF
MSMEG_3472|M.smegmatis_MC2_155 LRGGRPPLVIDSDGTVF
**** *** : ******