For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AKVEVQVASQMTPEQAWKLASDLGRFDEWLTIFGGWRSAVPDIIEVGTRVSALIKVKGFRNVIHWEVTQY DEPRRIEMRGHGRGGVRIALMMTVGEDPAGSCFHLVADLSGGLLSGPVGSLVAKVLTSDVRKSVQNLAAL H*
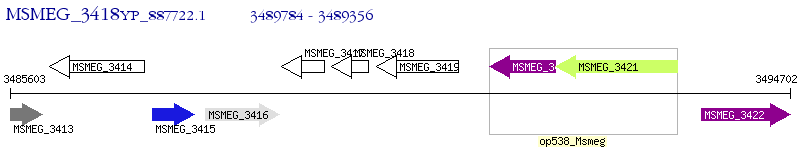
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3418 | - | - | 100% (142) | hypothetical protein MSMEG_3418 |
| M. smegmatis MC2 155 | MSMEG_5634 | - | 8e-28 | 38.03% (142) | hypothetical protein MSMEG_5634 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0934 | - | 1e-27 | 39.72% (141) | hypothetical protein Mb0934 |
| M. gilvum PYR-GCK | Mflv_3139 | - | 2e-48 | 65.96% (141) | hypothetical protein Mflv_3139 |
| M. tuberculosis H37Rv | Rv0910 | - | 1e-27 | 39.72% (141) | hypothetical protein Rv0910 |
| M. leprae Br4923 | MLBr_02113 | - | 8e-29 | 40.88% (137) | hypothetical protein MLBr_02113 |
| M. abscessus ATCC 19977 | MAB_0966 | - | 2e-27 | 39.55% (134) | hypothetical protein MAB_0966 |
| M. marinum M | MMAR_4619 | - | 3e-27 | 39.01% (141) | hypothetical protein MMAR_4619 |
| M. avium 104 | MAV_2938 | - | 9e-51 | 65.44% (136) | hypothetical protein MAV_2938 |
| M. thermoresistible (build 8) | TH_2177 | - | 1e-53 | 70.92% (141) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_1544 | - | 3e-27 | 37.32% (142) | hypothetical protein MUL_1544 |
| M. vanbaalenii PYR-1 | Mvan_3386 | - | 7e-47 | 65.49% (142) | hypothetical protein Mvan_3386 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3139|M.gilvum_PYR-GCK -------MATVDVSVTSDLSPEAAWELASDLKRFGEWLTIFAGWKSEVPA
Mvan_3386|M.vanbaalenii_PYR-1 -------MATVDVSVTSDLTPERAWVLASDLKRFDEWLTIFGGWKSAVPS
TH_2177|M.thermoresistible__bu -------MATVDVEVSSDLTPEQAWQRASDLRRFDDWLTIFGGWRSAVPS
MSMEG_3418|M.smegmatis_MC2_155 -------MAKVEVQVASQMTPEQAWKLASDLGRFDEWLTIFGGWRSAVPD
MAV_2938|M.avium_104 ------------MSVSSELEPETAWKLASDLDRFGEWMTIFAGWRGPVPD
Mb0934|M.bovis_AF2122/97 -------MAKLSGSIDVPLPPEEAWMHASDLTRYREWLTIHKVWRSKLPE
Rv0910|M.tuberculosis_H37Rv -------MAKLSGSIDVPLPPEEAWMHASDLTRYREWLTIHKVWRSKLPE
MLBr_02113|M.leprae_Br4923 MTSRIKRMAKLTGSIDVPLPPNEAWMCASDLARFGEWLTIHRAWRSKLPE
MMAR_4619|M.marinum_M -------MAKLSGSIDVPLSPDEAWMHASDLSRFKEWLTIHRVWRSTLPE
MAB_0966|M.abscessus_ATCC_1997 ------MAAKLNSSIDVPLPPEEAWAHASDLRRFDEWLSVHTAWRSALPE
MUL_1544|M.ulcerans_Agy99 -------MASVEMTADVPIPPQDTWDHVSDLSELGEWLVIHEGWRSELPE
: *: :* .*** . :*: :. *:. :*
Mflv_3139|M.gilvum_PYR-GCK EIEVDTQVSSLIKVKGFRNVIHWRVTRYDEPKEIELVGNGRGGVCIALTM
Mvan_3386|M.vanbaalenii_PYR-1 EIDVDTQVASLIKVKGFRNVIHWRVTRYDEPKEIELVGTGRGGVCIALSL
TH_2177|M.thermoresistible__bu EIEVGTCVSSLIKVKGFRNVIHWRVTRYDEPKEIELVGRGRGGVCIALTL
MSMEG_3418|M.smegmatis_MC2_155 IIEVGTRVSALIKVKGFRNVIHWEVTQYDEPRRIEMRGHGRGGVRIALMM
MAV_2938|M.avium_104 TIGEGTRVASCVKVKGFRNVIHWTVTRYDQPKSIELQGRGRGGIRLTVAM
Mb0934|M.bovis_AF2122/97 VLEKGTVVESYVEVKGMPNRIKWTIVRYKPPEGMTLNGDGVGGVKVKLIA
Rv0910|M.tuberculosis_H37Rv VLEKGTVVESYVEVKGMPNRIKWTIVRYKPPEGMTLNGDGVGGVKVKLIA
MLBr_02113|M.leprae_Br4923 VVEKGTVIESYVEVKGMPNRIRWTVVRYKAPEAMTLNGDGVGGVKVKLIA
MMAR_4619|M.marinum_M TLEKGAVVESIVQVKGMHNRIKWTIVRYQPPEGMTLNGDGVGGVKVKLMA
MAB_0966|M.abscessus_ATCC_1997 TLEKGTVIDSIVCVKGMYNRVKWTLQKYDPPTGLFLDGQGRGGVKVKLRV
MUL_1544|M.ulcerans_Agy99 QLSEGVEIEGVARAKGMRNRVTWTVTKWDPPREVTMSGSGKGGAKYGVSL
: .. : . .**: * : * : ::. * : : * * ** :
Mflv_3139|M.gilvum_PYR-GCK QVEPQ-GDGSSFGVIADLSGGLLSTPVGNVVARVIKGDVDRSVRNLAALS
Mvan_3386|M.vanbaalenii_PYR-1 RVEPAESGGSSFSVCADLFGGLLSTPVGHVVAKVIKGDVYRSVCNLAELR
TH_2177|M.thermoresistible__bu CVGQN-RSGTTFRVVADLSGGLLNTRIGKLVAKVLESDVRRSVTNLAALS
MSMEG_3418|M.smegmatis_MC2_155 TVGED-PAGSCFHLVADLSGGLLSGPVGSLVAKVLTSDVRKSVQNLAALH
MAV_2938|M.avium_104 TVTDD-HPGSVFHLTADIDGGVLSGPIGGLVARVLRSDVHKSVHNLAALQ
Mb0934|M.bovis_AF2122/97 KVAPK-EHGSVVSFDVHLGGPALLGPIGMIVAAALRADIRESLQNFVTVF
Rv0910|M.tuberculosis_H37Rv KVAPK-EHGSVVSFDVHLGGPALLGPIGMIVAAALRADIRESLQNFVTVF
MLBr_02113|M.leprae_Br4923 KVSPK-DDGSVVSFDVHLGGSALFGPIGMIVAVALRTDIRESLQNFVTAF
MMAR_4619|M.marinum_M KVAAG-EEGSIVSFDVHLGGPALFGPIGMVVAAALRSDINASLRNFVTVF
MAB_0966|M.abscessus_ATCC_1997 TISPK-GSGSVVDFNLHLGGPAMFGPIGAVVAAALRSDIDASLAKFVQVF
MUL_1544|M.ulcerans_Agy99 TVEPN-ADGSTLGLRLELGGGALFGPLGSAAARVVKGDVEQSLKQFVELY
: *: . . .: * : :* .* .: *: *: ::.
Mflv_3139|M.gilvum_PYR-GCK --------------------------------
Mvan_3386|M.vanbaalenii_PYR-1 --------------------------------
TH_2177|M.thermoresistible__bu --------------------------------
MSMEG_3418|M.smegmatis_MC2_155 --------------------------------
MAV_2938|M.avium_104 PSGPSAPGG-----------------------
Mb0934|M.bovis_AF2122/97 AG------------------------------
Rv0910|M.tuberculosis_H37Rv AG------------------------------
MLBr_02113|M.leprae_Br4923 SRPEPGLIRSRALVVDQHSRAVVEQRSASAGL
MMAR_4619|M.marinum_M APSAAG--------------------------
MAB_0966|M.abscessus_ATCC_1997 AATP----------------------------
MUL_1544|M.ulcerans_Agy99 G-------------------------------