For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSRGDFEALTNEPETALAPDQLPPREAVPAPQFDRLPGPQLLSYSAVTVVVALALLFGVGDAWRISLDRP TPPAPGEAQTAPGHKEMNLNARLRGMIEAVSNQFGPAAANNLLELRFTGSGSDYGTVLDPANGDTTIVYV NNGGDAFTTPSAQVLRKDSTFTAGDIASADLTAITRKMGERLDSAGGHVDLDSLEIERSRPGGPVILTGI FGSKTVKALPDGTVGEIFDPADFAVSFQRARESLTLAGITPSDRVLTNFEIRGTHRATPIAHASELQTRG GVLLEFQTEDRSGQIVIVPGELPEITQQAGHVGSQTFSFNDISPQVFDSVRAQAMQRGDLEPYEREAVHI WMTDRAIDPYRLAIRIELAGVEAATGTYSVTGEFLAPDAV
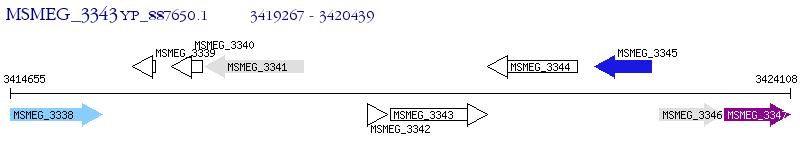
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3343 | - | - | 100% (390) | hypothetical protein MSMEG_3343 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2377 | - | 1e-138 | 64.34% (387) | hypothetical protein MAB_2377 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3343|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2377|M.abscessus_ATCC_1997 MNRATRRVGWVGAAGAVIGIGIGLGSYNPWPVALIVGIPLVTVGLAVVLR
MSMEG_3343|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2377|M.abscessus_ATCC_1997 VSRASEYERFRLAAASDGVPTRIEALTRSSVGDGDLQPTLMVATISPPHD
MSMEG_3343|M.smegmatis_MC2_155 ----------MSRGDFEALTNEPETALAPDQLPPREAVPAPQFDRLPGPQ
MAB_2377|M.abscessus_ATCC_1997 TEYQGRWIASMSGEDFKSLAASPVTALAPDRLPSRQVPGTPSFNDRRRTW
** **::*: .* ******:**.*:. :*.*: .
MSMEG_3343|M.smegmatis_MC2_155 LLSYSAVTVVVALALLFGVGDAWRISLDRPTPPAPGEAQTAPGHKEMNLN
MAB_2377|M.abscessus_ATCC_1997 ALISPALSILVAMAVLFGVGDAWHISLTTPAAPTP-SVNNDSKSKSMELN
* .*::::**:*:********:*** *:.*:* ..:. . *.*:**
MSMEG_3343|M.smegmatis_MC2_155 ARLRGMIEAVSNQFGPAAANNLLELRFTGSGSDYGTVLDPANGDTTIVYV
MAB_2377|M.abscessus_ATCC_1997 ARRDGMIRAITEQFGPAATNNLLDLRFTGSGSDYGTILDPTNGAVSIVYI
** ***.*:::******:****:************:***:** .:***:
MSMEG_3343|M.smegmatis_MC2_155 NNGGDAFTTPSAQVLRKDSTFTAGDIASADLTAITRKMGERLDSAGGHVD
MAB_2377|M.abscessus_ATCC_1997 NNNGDAFTTASPQVLRKDSTFTAGEIASTDLSALIEKMAHQADSAQRLSA
**.******.*.************:***:**:*: .**..: ***
MSMEG_3343|M.smegmatis_MC2_155 LDSLEIERSRPGGPVILTGIFGSKTVKALPDGTVGEIFDPADFAVSFQRA
MAB_2377|M.abscessus_ATCC_1997 LGTLEIKRSGPGAPVILTGSFGRRSINALPNGTVGEIFDPADFAVSFQRA
*.:***:** **.****** ** ::::***:*******************
MSMEG_3343|M.smegmatis_MC2_155 RESLTLAGITPSDRVLTNFEIRGTHRATPIAHASELQTRGGVLLEFQTED
MAB_2377|M.abscessus_ATCC_1997 RDALRLAGITASDRVLTDFEIRGSHRATPIARPSEVQNTGGVVLEFVTGD
*::* *****.******:*****:*******:.**:*. ***:*** * *
MSMEG_3343|M.smegmatis_MC2_155 RSGQIVIVPGELPEITQQAGHVGSQTFSFNDISPQVFDSVRAQAMQRGDL
MAB_2377|M.abscessus_ATCC_1997 RSGQIVLVPGEIPKVTDKSYRSASQPFAFDDVSAEVFESVRAQAMQRGAL
******:****:*::*::: : .**.*:*:*:*.:**:********** *
MSMEG_3343|M.smegmatis_MC2_155 EPYEREAVHIWMTDRAIDPYRLAIRIELAGVEAATGTYSVTGEFLAPDAV
MAB_2377|M.abscessus_ATCC_1997 EPYEREAVDIWVGDQVIDPFRLAIRVELAGVDAATGTYSLDGRFLAPGSR
********.**: *:.***:*****:*****:*******: *.****.: