For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTKRVVVLDYGSGNLRSAQRALERVGADVEVTADPSAAAAADGLVVPGVGAFEACMTGLRGIGGEKIIAD RLRNGAPVLGVCVGMQILFERGVEFGVETAGCGQWPGSVTRLDAPVIPHMGWNVVDAAKGSRLFAGLDAD TRFYFVHSYAAQQWSGREDALVTWATHHVPFIAAVEDGALSATQFHPEKSGDAGATLLANWVEGL
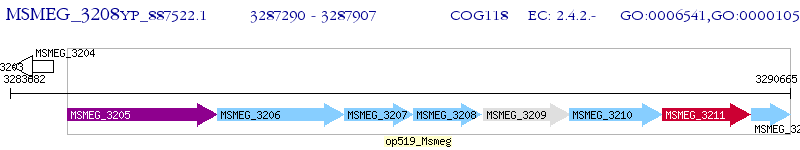
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3208 | hisH | - | 100% (205) | imidazole glycerol phosphate synthase subunit HisH |
| M. smegmatis MC2 155 | MSMEG_2939 | - | 9e-11 | 29.44% (180) | glutamine amidotransferase subunit PdxT |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1628 | hisH | 9e-92 | 77.83% (203) | imidazole glycerol phosphate synthase subunit HisH |
| M. gilvum PYR-GCK | Mflv_3610 | hisH | 2e-98 | 83.41% (205) | imidazole glycerol phosphate synthase subunit HisH |
| M. tuberculosis H37Rv | Rv1602 | hisH | 4e-91 | 77.34% (203) | imidazole glycerol phosphate synthase subunit HisH |
| M. leprae Br4923 | MLBr_01260 | hisH | 2e-90 | 78.11% (201) | imidazole glycerol phosphate synthase subunit HisH |
| M. abscessus ATCC 19977 | MAB_2667c | hisH | 3e-90 | 76.56% (209) | imidazole glycerol phosphate synthase subunit HisH |
| M. marinum M | MMAR_2398 | hisH | 3e-90 | 78.82% (203) | amidotransferase HisH |
| M. avium 104 | MAV_3184 | hisH | 9e-94 | 79.41% (204) | imidazole glycerol phosphate synthase subunit HisH |
| M. thermoresistible (build 8) | TH_1160 | hisH | 1e-95 | 80.30% (203) | Probable amidotransferase hisH |
| M. ulcerans Agy99 | MUL_1575 | hisH | 4e-89 | 78.61% (201) | imidazole glycerol phosphate synthase subunit HisH |
| M. vanbaalenii PYR-1 | Mvan_2807 | hisH | 4e-98 | 83.41% (205) | imidazole glycerol phosphate synthase subunit HisH |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2398|M.marinum_M MSGTPRPKSVVVLDYGSGNLRSAQRALQRVGADTVVTADEKAAMAADGLV
MUL_1575|M.ulcerans_Agy99 ---------MVVLDYGSGNLRSAQRALQRVGADTVVTADEKAAMAADGLV
Mb1628|M.bovis_AF2122/97 ----MTAKSVVVLDYGSGNLRSAQRALQRVGAEVEVTADTDAAMTADGLV
Rv1602|M.tuberculosis_H37Rv ----MTAKSVVVLDYGSGNLRSAQRALQRVGAEVEVTADTDAAMTADGLV
MLBr_01260|M.leprae_Br4923 ----MRSKSVVVLDYGSGNLWSVQRALQRVGAAVEVTADSAAGAAADGLL
MAV_3184|M.avium_104 ----MTSKSVVVLDYGSGNLRSAQRALERVGASVQVTADADAAAAADGLV
TH_1160|M.thermoresistible__bu VTAGSKRKTVAVLDYGSGNLRSAQRALERVGADVEVTADAQVALSADGLV
Mflv_3610|M.gilvum_PYR-GCK -----MTKKLVVLDYGSGNLRSAQRALERVGAEVEVTADSDAAANADGLV
Mvan_2807|M.vanbaalenii_PYR-1 -----MTTKLVVLDYGSGNLRSAQRALERVGAHVEVTADAEAAANADGLV
MSMEG_3208|M.smegmatis_MC2_155 -----MTKRVVVLDYGSGNLRSAQRALERVGADVEVTADPSAAAAADGLV
MAB_2667c|M.abscessus_ATCC_199 --MTGAGPKVVVLDYGSGNLRSAQRALERVGADVTVTADSSTALNADGLV
:.********* *.****:**** . **** .. ****:
MMAR_2398|M.marinum_M VPGVGAFEACMTGLRKIGGERIIAARVAAGRPVLGVCVGMQILFARGVEF
MUL_1575|M.ulcerans_Agy99 VPGVGAFEACMTGLRKIGGERIIAARVAAGRPVLGVCVGMQILFARGVEF
Mb1628|M.bovis_AF2122/97 VPGVGAFAACMAGLRKISGERIIAERVAAGRPVLGVCVGMQILFACGVEF
Rv1602|M.tuberculosis_H37Rv VPGVGAFAACMAGLRKISGERIIAERVAAGRPVLGVCVGMQILFACGVEF
MLBr_01260|M.leprae_Br4923 VPGVGAFEACMAGLRKIAGERTIAERIVAGRPVLGVCVGMQILFARGVEF
MAV_3184|M.avium_104 VPGVGAYEACMTGLRKIGGDRIIAERVAAGRPVLGVCVGMQILFARGVEF
TH_1160|M.thermoresistible__bu VPGVGAFAACMSGLHAVGGDRIIAERVAAGRPVLGVCVGMQILFARGVEF
Mflv_3610|M.gilvum_PYR-GCK VPGVGAFEACMTGLRGIGGEKIIADRLAAGRPVLGVCVGMQILFARGVEF
Mvan_2807|M.vanbaalenii_PYR-1 VPGVGAFQACMTGLRAIGGDKIIADRVSAGRPVLGVCVGMQIMFARGVEF
MSMEG_3208|M.smegmatis_MC2_155 VPGVGAFEACMTGLRGIGGEKIIADRLRNGAPVLGVCVGMQILFERGVEF
MAB_2667c|M.abscessus_ATCC_199 VPGVGAFAACMEGLRGIDGERIIDIRLSGGRPVLGICVGMQILFSHGIEF
******: *** **: : *:: * *: * ****:******:* *:**
MMAR_2398|M.marinum_M GVETQGCGLWPGAVTRLDAPVIPHMGWNVVTAPADSVLFKGLEPKARFYF
MUL_1575|M.ulcerans_Agy99 GVETQGCGLWPGAVTRLDAPVIPHMGWNVVTAPADSVLFKGLEPKARFYF
Mb1628|M.bovis_AF2122/97 GVQTPGCGHWPGAVIRLEAPVIPHMGWNVVDSAAGSALFKGLDVDARFYF
Rv1602|M.tuberculosis_H37Rv GVQTPGCGHWPGAVIRLEAPVIPHMGWNVVDSAAGSALFKGLDVDARFYF
MLBr_01260|M.leprae_Br4923 GVETTGCRQWPGVVTRLDAPVVPHMGWNVVDSASGSALFKGLDAGVRFYF
MAV_3184|M.avium_104 SVETSGCGQWPGSVTRLQAPVIPHMGWNVVESGPGSVLFRGLDADTRFYF
TH_1160|M.thermoresistible__bu GVESRGCGQWPGSVVRLDAPVVPHLGWNVVDAPAGSRLFAGLDSCTRFYF
Mflv_3610|M.gilvum_PYR-GCK GVESTGCGQWPGAVVRLDAPVVPHMGWNVVDAPADSVLFKGMDAETRFYF
Mvan_2807|M.vanbaalenii_PYR-1 GVESAGCGQWPGSVIRLDAPVIPHMGWNVVDAPAGSVLFKGIDADTRFYF
MSMEG_3208|M.smegmatis_MC2_155 GVETAGCGQWPGSVTRLDAPVIPHMGWNVVDAAKGSRLFAGLDADTRFYF
MAB_2667c|M.abscessus_ATCC_199 GVDTQGCGQWPGVVSRLDAPVIPHMGWNTVDAAPGSQLFAGLDAGTRFYF
.*:: ** *** * **:***:**:***.* : .* ** *:: .****
MMAR_2398|M.marinum_M VHSYAAQRWEGSSG-------ARLTWATHQVPFLAAVEDGPLAATQFHPE
MUL_1575|M.ulcerans_Agy99 VHSYAAQRWEGSSG-------ARLTWATHQVSFLAAVEDGPLAATQFHPE
Mb1628|M.bovis_AF2122/97 VHSYAAQRWEGSPD-------ALLTWATYRAPFLAAVEDGALAATQFHPE
Rv1602|M.tuberculosis_H37Rv VHSYAAQRWEGSPD-------ALLTWATYRAPFLAAVEDGALAATQFHPE
MLBr_01260|M.leprae_Br4923 VHSYAAQRWEGSSK-------ALLTWATHQVPFLAAVEEGPLVATQFHPE
MAV_3184|M.avium_104 VHSYAAQQWEGSPE-------AVLTWSRHEGPFLAAVEDGPLSATQFHPE
TH_1160|M.thermoresistible__bu VHSYAAQQWEGDPA-------ALLTWSTHHVPFLAAVEDGPLAATQFHPE
Mflv_3610|M.gilvum_PYR-GCK VHSYAAQQWEGDPS-------ALLTWATHHVPFLAAVENGPLSATQFHPE
Mvan_2807|M.vanbaalenii_PYR-1 VHSYAAQQWEGGSG-------ALLTWATHHVPFLAAVEDGPLSATQFHPE
MSMEG_3208|M.smegmatis_MC2_155 VHSYAAQQWSGRED-------ALVTWATHHVPFIAAVEDGALSATQFHPE
MAB_2667c|M.abscessus_ATCC_199 VHSYAVQKWELETPGESPIAPPLVTWATHHVPFVAAVENGPLAATQFHPE
*****.*:*. . :**: :. .*:****:*.* *******
MMAR_2398|M.marinum_M KSGDAGAAVLSNWVENL-
MUL_1575|M.ulcerans_Agy99 KSGDAGAAVLSNWVENL-
Mb1628|M.bovis_AF2122/97 KSGDAGAAVLSNWVDGL-
Rv1602|M.tuberculosis_H37Rv KSGDAGAAVLSSWVDGL-
MLBr_01260|M.leprae_Br4923 KSGDAGATLLSNWLGEL-
MAV_3184|M.avium_104 KSGDAGAAVLRNWIERL-
TH_1160|M.thermoresistible__bu KSGDAGATLLANWVAGI-
Mflv_3610|M.gilvum_PYR-GCK KSGDAGAELLSNWVGALS
Mvan_2807|M.vanbaalenii_PYR-1 KSGDAGAELLSNWVGALS
MSMEG_3208|M.smegmatis_MC2_155 KSGDAGATLLANWVEGL-
MAB_2667c|M.abscessus_ATCC_199 KSGDAGAVLLSNWVKGIS
******* :* .*: :