For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAVVFQVRDLDTRQPSLNVLLWQRALEPERDKWSLPGGRLRDDEDLTTSVRRQLAEKVDLRELAHLEQLA VFSDPKRVPGERTIASTFLGLVPSPATPALPDDTRWHPVHELPPMAFDHAPMVEHARTRLVAKLSYTNIG FALAPKEFAISSLRDVYSAALDYQVDATNLQRVLERRKVITRTGTTARSGRSGGRPAALFRFTESRYRVT DEFAALRPPG*
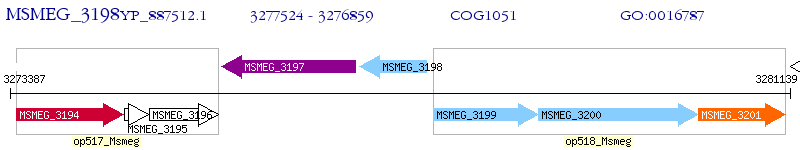
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3198 | - | - | 100% (221) | hydrolase, NUDIX family protein |
| M. smegmatis MC2 155 | MSMEG_1268 | - | 8e-13 | 30.59% (170) | nudix hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1619c | - | 1e-101 | 79.56% (225) | hypothetical protein Mb1619c |
| M. gilvum PYR-GCK | Mflv_3620 | - | 1e-104 | 83.11% (225) | NUDIX hydrolase |
| M. tuberculosis H37Rv | Rv1593c | - | 1e-101 | 79.56% (225) | hypothetical protein Rv1593c |
| M. leprae Br4923 | MLBr_01224 | - | 3e-96 | 75.11% (229) | hypothetical protein MLBr_01224 |
| M. abscessus ATCC 19977 | MAB_2675 | - | 2e-96 | 77.93% (222) | hypothetical protein MAB_2675 |
| M. marinum M | MMAR_2390 | - | 6e-98 | 77.88% (226) | hypothetical protein MMAR_2390 |
| M. avium 104 | MAV_3192 | - | 5e-96 | 76.89% (225) | hydrolase, NUDIX family protein |
| M. thermoresistible (build 8) | TH_0810 | - | 1e-106 | 84.09% (220) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1567 | - | 6e-98 | 77.88% (226) | hypothetical protein MUL_1567 |
| M. vanbaalenii PYR-1 | Mvan_2797 | - | 1e-103 | 81.78% (225) | NUDIX hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3620|M.gilvum_PYR-GCK MAHTSTDHEVLAVVFQVRRAESPTA----KRKPALHVLLWQRALEPEKEK
Mvan_2797|M.vanbaalenii_PYR-1 MAHISTEHEVLAAVFQVRPGRPPVT----SRKPALHVLLWERALDPEKGR
TH_0810|M.thermoresistible__bu ---------VLAVVFQVRG----LD----SRQPALHVLLWQRALEPERGR
MSMEG_3198|M.smegmatis_MC2_155 ---------MLAVVFQVRD----LD----TRQPSLNVLLWQRALEPERDK
Mb1619c|M.bovis_AF2122/97 MAHGSTAHEVLAVVFQVRGVGMSRG----AAKPQLNVLLWQRAKEPQRGA
Rv1593c|M.tuberculosis_H37Rv MAHGSTAHEVLAVVFQVRGVGMSRG----AAKPQLNVLLWQRAKEPQRGA
MAV_3192|M.avium_104 ---------MFQVREASEPVTKGRH----RAKPQLNVLLWQRAKEPQRGA
MLBr_01224|M.leprae_Br4923 MAYASTAHEVLAVVFQVRRVAEQVKGTPQKAKPQLSVLLWERSQDPQRGA
MMAR_2390|M.marinum_M MPNSNTAHEVLAAVFQVRQITAASR----GGKPQLNVLLWQRAQDPQKGA
MUL_1567|M.ulcerans_Agy99 MPNSNTAHEVLAAVFQVRHITAASR----RGKPQLNVLLWQRAQDPQKGA
MAB_2675|M.abscessus_ATCC_1997 MVHTSTEHEVLAVVFQVRHFDKAQT-------PELAVLLWQRALDPHRGA
:: . . * * ****:*: :*.:
Mflv_3620|M.gilvum_PYR-GCK WALPGGGLGADEDLTSSVRRQLAEKVDLRELAHLEQLAIFSDPHRVPG-P
Mvan_2797|M.vanbaalenii_PYR-1 WALPGGHLGPDEDLTSSVRRQLAEKVDLRELAHLEQLAVFSDPERVPG-P
TH_0810|M.thermoresistible__bu WSLPGGLLGTDEDLTSSVRRQLAEKVDLRELAHLEQLAVFSDPHRVPG-V
MSMEG_3198|M.smegmatis_MC2_155 WSLPGGRLRDDEDLTTSVRRQLAEKVDLRELAHLEQLAVFSDPKRVPG-E
Mb1619c|M.bovis_AF2122/97 WSLPGGRLRNDEDMTSSVRRQLAEKVDLRELAHLEQLAVFSDPHRLPG-I
Rv1593c|M.tuberculosis_H37Rv WSLPGGRLRNDEDMTSSVRRQLAEKVDLRELAHLEQLAVFSDPHRLPG-I
MAV_3192|M.avium_104 WSLPGGRLRNDEDMTTSVRRQLAEKVDLKELAHLEQLAVFSDPNRVPG-T
MLBr_01224|M.leprae_Br4923 WSLPGGRLRNDEDMTYSVRRQLAEKVDLRELAHLEQLAVFSEPARLPG-T
MMAR_2390|M.marinum_M WSLPGGRLRSDEDLTTSVRRQLAEKVDLRELSHLEQLAVFSDPHRVPGDT
MUL_1567|M.ulcerans_Agy99 WSLPGGRLRSDEDLTTSVRRQLAEKVDLRELSHLEQLAVFSDPHRVPGDT
MAB_2675|M.abscessus_ATCC_1997 WALPGGRVRADEDLPSSIRRQLAEKVDVRELAHLEQLAVFSEPRRVPG-P
*:**** : ***:. *:*********::**:******:**:* *:**
Mflv_3620|M.gilvum_PYR-GCK RTIASTFLGLVPSPATPELPPDTRWHPVSDLPPMAFDHGPMVEHARTRLE
Mvan_2797|M.vanbaalenii_PYR-1 RTIASTFLGLVPSPATPELPPDTRWHPVSELPAMAFDHGPMVEHAHNRLV
TH_0810|M.thermoresistible__bu RTIASTYLGLVPSPASPALPDDTRWHPVHALPPMAFDHGPMVEHARSRLV
MSMEG_3198|M.smegmatis_MC2_155 RTIASTFLGLVPSPATPALPDDTRWHPVHELPPMAFDHAPMVEHARTRLV
Mb1619c|M.bovis_AF2122/97 RMIASTYLGVVPSPATPELPADTRWHPVSSLPPMAFDHGPMVTHARTRLI
Rv1593c|M.tuberculosis_H37Rv RMIASTYLGVVPSPATPELPADTRWHPVSSLPPMAFDHGPMVTHARTRLI
MAV_3192|M.avium_104 RMIASTFLGLVPSPASPELPPDTRWHPVNCLPPMAFDHGQMVDNARARLI
MLBr_01224|M.leprae_Br4923 RMIASTFLGLVPSPATPELPPDTRWHPLNTLPSMAFDHGPMVTHARARLV
MMAR_2390|M.marinum_M RVIASTFLGLVPSPATPELPADTRWHPVNALPPMAFDHGPMVTHAHARLV
MUL_1567|M.ulcerans_Agy99 RVIASTFLGLVPSPATPELPADTRWHPVNALPPMAFDHGPMVTHAHARLV
MAB_2675|M.abscessus_ATCC_1997 RTIASTFLGLVPFPATPALPDDTQWHRVNHLPAMAFDHQVMVAHAHTRLA
* ****:**:** **:* ** **:** : **.***** ** :*: **
Mflv_3620|M.gilvum_PYR-GCK AKLSYTNIGFALAPNEFALSALRDIYSAALGHPVDATNLQRVLERRHVIT
Mvan_2797|M.vanbaalenii_PYR-1 AKLSYTNIGFALAPSEFALSTLRDIYSAALGHPVDATNLQRVLERRRVIT
TH_0810|M.thermoresistible__bu AKLSYTNIGFGLAPEEFALSTLRDIYAAALGHPVDTTNLQRVLERRHVIT
MSMEG_3198|M.smegmatis_MC2_155 AKLSYTNIGFALAPKEFAISSLRDVYSAALDYQVDATNLQRVLERRKVIT
Mb1619c|M.bovis_AF2122/97 AKMSYTNIGFALAPKEFALSTLRDIYGAALGYQVDATNLQRVLARRRVIT
Rv1593c|M.tuberculosis_H37Rv AKMSYTNIGFALAPKEFALSTLRDIYGAALGYQVDATNLQRVLARRRVIT
MAV_3192|M.avium_104 AKLSYTNIGFALAPREFALSTLRDIYGAALGYQVDATNLQRVLARRGVIT
MLBr_01224|M.leprae_Br4923 AKMSYTNIGFALAPKEFALSTLRDIYGATLGYQVDATNLQRVLARRSVII
MMAR_2390|M.marinum_M AKMSYTNIGFALTPKEFALSTLRDIYGATLGYQVDTTNLQRVLARRKVIT
MUL_1567|M.ulcerans_Agy99 AKMSYTNIGFALTPKEFALSTLRDIYGATLGYQVDTTNLQRVLARRKVIT
MAB_2675|M.abscessus_ATCC_1997 AKMSYTNIGFALAPQEFTLSTLRDIYCATLGYPVDATNLQRVLVRRGVLT
**:*******.*:* **::*:***:* *:*.: **:******* ** *:
Mflv_3620|M.gilvum_PYR-GCK RTGTTARSGRSGGRPAALYRFTEAAYRVTDEFAALRPPG---
Mvan_2797|M.vanbaalenii_PYR-1 RTGTTARSGRSGGRPAALYRFTDSRYRVTDEFAALRPPG---
TH_0810|M.thermoresistible__bu RTGSTARSGRAGGRPPALYRFTENRYRVTDEYAAFRPPAV--
MSMEG_3198|M.smegmatis_MC2_155 RTGTTARSGRSGGRPAALFRFTESRYRVTDEFAALRPPG---
Mb1619c|M.bovis_AF2122/97 QTGTIAQSGRSGGRPAALYRFTDSQLRVTDEFAALRPPGQL-
Rv1593c|M.tuberculosis_H37Rv QTGTIAQSGRSGGRPAALYRFTDSQLRVTDEFAALRPPGQL-
MAV_3192|M.avium_104 ATGTIAQSGRSGGRPAALYRFTDAALRVTDEFAALRPPGLPG
MLBr_01224|M.leprae_Br4923 QTGTVAQSGRSGGRPAALYRFTDSQLRVTDEFAALRPPGN--
MMAR_2390|M.marinum_M QTGTFAQSGRSGGRPAAMYRFTDSQLRVTDEFAALRPPG---
MUL_1567|M.ulcerans_Agy99 QTGTFAQSGRSGGRPAAMYRFTDSQLRVTDEFAALRPPG---
MAB_2675|M.abscessus_ATCC_1997 PTGTTARSGRSGGRPAALYRFTDDRYRVTDEFAALRPPG---
**: *:***:****.*::***: *****:**:***.