For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VESHEGIDVVLVTGLSGAGRGTAAKVLEDLGWYVADNLPPELIARMVDLGLAAGSRITQLAVVMDVRSKG FTGDLDWVRNELATRNIAPRVLFMEASDDILVRRYEQNRRSHPLQGTQTLAEGIAAERAMLAPVRAAADL VIDTSTLPVPALRESIERAFGGETVAYTNVTVESFGYKYGLPMDADTVMDVRFLPNPHWVDELRPHSGQH PDVRDYVLGQPGALEFLDTYHRLLDVVIDGYRREGKRYMTVAIGCTGGKHRSVAIAEALAERLEGGDGLT VRVLHRDLGRE
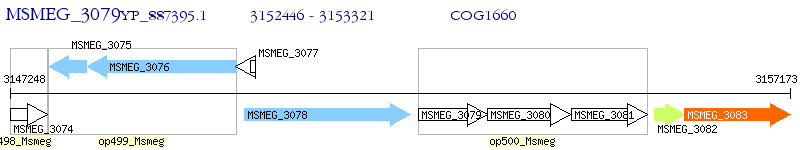
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3079 | - | - | 100% (291) | hypothetical protein MSMEG_3079 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1456 | - | 1e-135 | 83.92% (286) | hypothetical protein Mb1456 |
| M. gilvum PYR-GCK | Mflv_3714 | - | 1e-147 | 88.28% (290) | hypothetical protein Mflv_3714 |
| M. tuberculosis H37Rv | Rv1421 | - | 1e-135 | 83.92% (286) | hypothetical protein Rv1421 |
| M. leprae Br4923 | MLBr_00563 | - | 1e-134 | 84.56% (285) | hypothetical protein MLBr_00563 |
| M. abscessus ATCC 19977 | MAB_2783c | - | 1e-135 | 82.81% (285) | hypothetical protein MAB_2783c |
| M. marinum M | MMAR_2228 | - | 1e-135 | 82.41% (290) | hypothetical protein MMAR_2228 |
| M. avium 104 | MAV_3359 | - | 1e-138 | 83.67% (294) | hypothetical protein MAV_3359 |
| M. thermoresistible (build 8) | TH_1324 | - | 1e-149 | 90.66% (289) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1815 | - | 1e-135 | 82.41% (290) | hypothetical protein MUL_1815 |
| M. vanbaalenii PYR-1 | Mvan_2698 | - | 1e-147 | 90.91% (286) | hypothetical protein Mvan_2698 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3079|M.smegmatis_MC2_155 ------------------------------MES--------HEGIDVVLV
TH_1324|M.thermoresistible__bu MTDYHSDDELRGGPGVG-----------ATLDSGTDPEAGTESGIDVVLV
Mflv_3714|M.gilvum_PYR-GCK MTRQGMRDDLRGEADSVVHDGTDDIDNENDIDNGIDNE--NGNGIDVVLV
Mvan_2698|M.vanbaalenii_PYR-1 MTEQGMHQELREGAGTAGDEGG---LEAAAMDGGADN------GIDVVLV
MMAR_2228|M.marinum_M MTNH-----------------------IAGGGNPPGPEGDASAGIDVVLV
MUL_1815|M.ulcerans_Agy99 MTNH-----------------------IAGGGNPPGPEGDASAGIDVVLV
MLBr_00563|M.leprae_Br4923 MS----------------------------GNNHPG---DASAEIDVVLV
Mb1456|M.bovis_AF2122/97 MMNH-----------------------ARGVENRSE-----GGGIDVVLV
Rv1421|M.tuberculosis_H37Rv MMNH-----------------------ARGVENRSE-----GGGIDVVLV
MAV_3359|M.avium_104 ------------------------------MDAHLEN----GAGIDVVLV
MAB_2783c|M.abscessus_ATCC_199 ---------------------------MESMTLHPPSE-TANADIDVVLV
******
MSMEG_3079|M.smegmatis_MC2_155 TGLSGAGRGTAAKVLEDLGWYVADNLPPELIARMVDLGLAAGSRITQLAV
TH_1324|M.thermoresistible__bu TGLSGAGRGTAAKVLEDLGWYVADNLPPELIARMVELGLAAGSRITQLAV
Mflv_3714|M.gilvum_PYR-GCK TGLSGAGRGTAAKVLEDLGWYVADNLPPELIAQMVDLGLAAGSRITQLAA
Mvan_2698|M.vanbaalenii_PYR-1 TGLSGAGRGTAAKVLEDLGWYVADNLPPELIAQMVDLGLAAGSRITQLAV
MMAR_2228|M.marinum_M TGLSGAGRGTAAKVLEDLGWYVADNLPPQLITRMVDFGLAAGSRITQLAV
MUL_1815|M.ulcerans_Agy99 TGLSGAGRGTAAKVLEDLGWYVADNLPPQLITRMVDFGLAAGSRITQLAV
MLBr_00563|M.leprae_Br4923 TGLSGAGRGTAAKVLEDLGWYVADNLPPQLITWMVDFGLAAGSRITQLAV
Mb1456|M.bovis_AF2122/97 TGLSGAGRGTAAKVLEDLGWYVADNLPPQLITRMVDFGLAAGSRITQLAV
Rv1421|M.tuberculosis_H37Rv TGLSGAGRGTAAKVLEDLGWYVADNLPPQLITRMVDFGLAAGSRITQLAV
MAV_3359|M.avium_104 TGLSGAGRGTAAKVLEDLGWYVADNLPPQLITRMVDLGMDAGSRITQLAV
MAB_2783c|M.abscessus_ATCC_199 TGLSGAGRGTTAKVLEDLGWYVADNLPPELITRMVDLGLAAGSRITQLAV
**********:*****************:**: **::*: *********.
MSMEG_3079|M.smegmatis_MC2_155 VMDVRSKGFTGDLDWVRNELATRNIAPRVLFMEASDDILVRRYEQNRRSH
TH_1324|M.thermoresistible__bu VMDVRSRGFTGDLDGVRQELATRNIVPRVLFLDASDDILVRRYEQNRRSH
Mflv_3714|M.gilvum_PYR-GCK VMDVRSRGFTGDLDWVRSELATRNIVPRVVFLEASDDILVRRYEQNRRSH
Mvan_2698|M.vanbaalenii_PYR-1 VMDVRSRGFTGDLDWVRTELATRNIVPRVLFLEASDDILVRRYEQNRRSH
MMAR_2228|M.marinum_M VMDVRSRGFTGDLDSVRNELATRNITPRVVFMEASDDMLVRRYEQNRRSH
MUL_1815|M.ulcerans_Agy99 VMDVRSRGFTGDLDSVRNELATRNITPRVVFMEASDDMLVRRYEQNRRSH
MLBr_00563|M.leprae_Br4923 VMDVRSRGFTGDLDSVRRELATRNIIPRVVFMEASDDMLVRRYEQNRRSH
Mb1456|M.bovis_AF2122/97 VMDVRSRGFTGDLDSVRNELATRAITPRVVFMEASDDTLVRRYEQNRRSH
Rv1421|M.tuberculosis_H37Rv VMDVRSRGFTGDLDSVRNELATRAITPRVVFMEASDDTLVRRYEQNRRSH
MAV_3359|M.avium_104 VMDVRSRGFTGDLDSVRTELATRNIAPRVVFMEASDDMLVRRYEQNRRSH
MAB_2783c|M.abscessus_ATCC_199 VMDVRSRGFTGDLESVRADLATRGISPRVLFLEASDESLVRRYENNRRSH
******:******: ** :**** * ***:*::***: ******:*****
MSMEG_3079|M.smegmatis_MC2_155 PLQGTQTLAEGIAAERAMLAPVRAAADLVIDTSTLPVPALRESIERAFGG
TH_1324|M.thermoresistible__bu PLQGRQTLAEGIAAERAMLAPVRAAADLVIDTSALSVPALRENIERAFGG
Mflv_3714|M.gilvum_PYR-GCK PLQGNQTLAEGIAAERAMLAPVRASADLVIDTSALSVHGLRDSIERAFAA
Mvan_2698|M.vanbaalenii_PYR-1 PLQGNQTLAEGIAAERTMLAPVRASADLVIDTSSLSVHALRDSIERAFAG
MMAR_2228|M.marinum_M PLQGGHTLAEGIAAERKMLEPVRATADLIIDTSTLSVRGLRENIERAFGG
MUL_1815|M.ulcerans_Agy99 PLQGGHTLAEGIAAERKMLEPVRATADLIIDTSTLSVRGLRENIERAFGG
MLBr_00563|M.leprae_Br4923 PLQGEQTLAEGIAAERRMLAPVRATADLIIDTSALSVPGLRESIERAFGG
Mb1456|M.bovis_AF2122/97 PLQGEQTLAEGIAAERRMLAPVRATADLIIDTSTLSVGGLRDSIERAFGG
Rv1421|M.tuberculosis_H37Rv PLQGEQTLAEGIAAERRMLAPVRATADLIIDTSTLSVGGLRDSIERAFGG
MAV_3359|M.avium_104 PLQGDQTLAEGIAAERRMLAPVRATADLIIDTSTLSVRALRETIERAFGG
MAB_2783c|M.abscessus_ATCC_199 PLQGGQTLAEGIAAERALLSSIRASADLVIDTSSLPVPALRAAIERAFSS
**** :********** :* .:**:***:****:*.* .** *****..
MSMEG_3079|M.smegmatis_MC2_155 ETVAYTNVTVESFGYKYGLPMDADTVMDVRFLPNPHWVDELRPHSGQHPD
TH_1324|M.thermoresistible__bu DTVAHTNVTVESFGYKYGLPMDADTVMDVRFLPNPHWVDALRPYSGQHEA
Mflv_3714|M.gilvum_PYR-GCK ETVAHTNVTVESFGYKYGLPMDADTVMDVRFLPNPHWIDRLRPHTGQNAD
Mvan_2698|M.vanbaalenii_PYR-1 ETVAHTNVTVESFGYKYGLPMDADTVMDVRFLPNPHWVDRLRPNTGQHHE
MMAR_2228|M.marinum_M DAAAITSVTVESFGFKYGLPMDADMVMDVRFLPNPHWVDELRPLTGQHQA
MUL_1815|M.ulcerans_Agy99 DAAAITSVTVESFGFKYGLPMDADMVMDVRFLPNPHWVDELRPLTGQHQA
MLBr_00563|M.leprae_Br4923 DASATTSVTVESFGFKYGLPMDADIVMDVRFLPNPHWVDELRSLTGQHSA
Mb1456|M.bovis_AF2122/97 DGGATTSVTVESFGFKYGLPMDADMVMDVRFLPNPHWVDELRPLTGQHPA
Rv1421|M.tuberculosis_H37Rv DGGATTSVTVESFGFKYGLPMDADMVMDVRFLPNPHWVDELRPLTGQHPA
MAV_3359|M.avium_104 GASATISVTVESFGFKYGLPMDADMVMDVRFLPNPHWVDELRPRTGQDPA
MAB_2783c|M.abscessus_ATCC_199 ESVAHISVTVESFGFKYGLPMDADMVVDVRFLPNPHWVDELRPHTGQHPS
* .*******:********* *:**********:* **. :**.
MSMEG_3079|M.smegmatis_MC2_155 VRDYVLGQPGALEFLDTYHRLLDVVIDGYRREGKRYMTVAIGCTGGKHRS
TH_1324|M.thermoresistible__bu VRDYVLSQPGAADFLDTYHRLLNLVIDGYRREGKRYMTVAIGCTGGKHRS
Mflv_3714|M.gilvum_PYR-GCK VRDYVLGQPGADEFLDSYHQLLNVVIDGYRREGKRYMTVAIGCTGGKHRS
Mvan_2698|M.vanbaalenii_PYR-1 VRDYVLGQPGALEFLDSYHQLLDVVIDGYRREGKRYMTVAIGCTGGKHRS
MMAR_2228|M.marinum_M VSDYVLGRPGASEFLQTYHELLSLVVEGYRREGKRYMTVAIGCTGGKHRS
MUL_1815|M.ulcerans_Agy99 VSDYVLGRPGASEFLQTYHELLSLVVEGYRREGKRYMTVAIGCTGGKHRS
MLBr_00563|M.leprae_Br4923 VRDYVLGQPGAAEFLRTYRRLLSLVVDGYRREGKRYMTVAIGCTGGKHRS
Mb1456|M.bovis_AF2122/97 VRDYVLHRPGAAEFLESYHRLLSLVVDGYRREGKRYMTIAIGCTGGKHRS
Rv1421|M.tuberculosis_H37Rv VRDYVLHRPGAAEFLESYHRLLSLVVDGYRREGKRYMTIAIGCTGGKHRS
MAV_3359|M.avium_104 VRDYVLGQPGAAEFLDAYHRLLSLVVDGYRREGKRYMTIAIGCTGGKHRS
MAB_2783c|M.abscessus_ATCC_199 VSHYVLSQPGADEFLDTYHRLLNLVIDGYRREGKRYMTIAVGCTGGKHRS
* .*** :*** :** :*:.**.:*::***********:*:*********
MSMEG_3079|M.smegmatis_MC2_155 VAIAEALAERLEGGDG---LTVRVLHRDLGRE
TH_1324|M.thermoresistible__bu VAIAEALAARLEGGDQ---LTVRVLHRDLGRE
Mflv_3714|M.gilvum_PYR-GCK VAMAEALAARLAGSDA---LTVRVLHRDLGRE
Mvan_2698|M.vanbaalenii_PYR-1 VAMAEALAERLQDSD----LTVRVLHRDLGRE
MMAR_2228|M.marinum_M VAIAEALVGLLRSNS---RLSVRVLHRDLGRE
MUL_1815|M.ulcerans_Agy99 VAIAEALVGLLRSNS---RLSVRVLHRDLGRE
MLBr_00563|M.leprae_Br4923 VAIAEALMGLLQSDL---QLSVRVLHRDLGRE
Mb1456|M.bovis_AF2122/97 VAIAEALMGLLRSDQ---QLSVRALHRDLGRE
Rv1421|M.tuberculosis_H37Rv VAIAEALMGLLRSDQ---QLSVRALHRDLGRE
MAV_3359|M.avium_104 VAIAEALMQRLQAQHGEAQLSVRVLHRDLGRE
MAB_2783c|M.abscessus_ATCC_199 VAITEALAASLAPDDD---LSVRVLHRDLGRE
**::*** * *:**.********