For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAIEEILDLEQLEVNIYRGRVFSPESGFLQRTFGGHVAGQSLVSAVRTVAPEFEVHSLHGYFLRPGDARA PAIYTVERIRDGGSFCTRRVSAIQHGETIFSMSASFQTDQSGIEHQDAMPHAPAPEECPGFRSGGAFDDA GFAQFAEWDVRIVPRDRVQKIPGKASQQQVWFRHRDPMPDDHVLHICALAYMSDLTLLGSAQVNHLDVRK HLMVASLDHAMWFMRPFRADEWLLYDQTSPSACGGRALTQGRIFNQYGELVAAVMQEGLTRYKRDFTPPE ENS
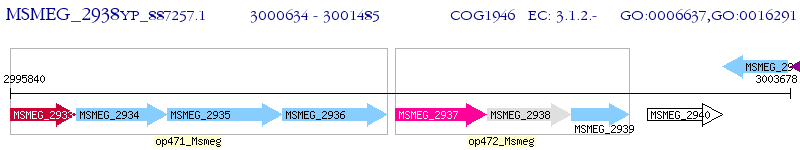
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2938 | tesB | - | 100% (283) | acyl-CoA thioesterase II |
| M. smegmatis MC2 155 | MSMEG_3228 | - | 1e-36 | 33.09% (278) | acyl-CoA thioesterase II |
| M. smegmatis MC2 155 | MSMEG_6208 | - | 9e-34 | 36.00% (250) | palmitoyl-CoA hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2637c | tesB2 | 1e-128 | 79.56% (274) | acyl-CoA thioesterase II |
| M. gilvum PYR-GCK | Mflv_3831 | - | 1e-140 | 84.59% (279) | acyl-CoA thioesterase II |
| M. tuberculosis H37Rv | Rv2605c | tesB2 | 1e-128 | 79.93% (274) | acyl-CoA thioesterase II |
| M. leprae Br4923 | MLBr_01278 | tesB | 7e-30 | 31.72% (290) | acyl CoA thioesterase II |
| M. abscessus ATCC 19977 | MAB_2891c | - | 3e-96 | 64.41% (281) | acyl-CoA thioesterase II TesB2 |
| M. marinum M | MMAR_2095 | tesB2 | 1e-128 | 79.00% (281) | acyl-CoA thioesterase II TesB2 |
| M. avium 104 | MAV_3483 | tesB | 1e-126 | 77.90% (276) | acyl-CoA thioesterase II |
| M. thermoresistible (build 8) | TH_2056 | tesB2 | 1e-143 | 85.41% (281) | PROBABLE ACYL-CoA THIOESTERASE II TESB2 (TEII) |
| M. ulcerans Agy99 | MUL_3249 | tesB2 | 1e-128 | 79.29% (280) | acyl-CoA thioesterase II TesB2 |
| M. vanbaalenii PYR-1 | Mvan_2565 | - | 1e-139 | 84.59% (279) | acyl-CoA thioesterase II |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2637c|M.bovis_AF2122/97 ----------MSIEEILDLEQLEVNIYRGSVFSPESGFLQRTFGGHVAGQ
Rv2605c|M.tuberculosis_H37Rv ----------MSIEEILDLEQLEVNIYRGSVFSPESGFLQRTFGGHVAGQ
MAV_3483|M.avium_104 ----------MSIEQILDLEQLEVNIYRGGVFSPDSGYFQRTFGGHVAGQ
MMAR_2095|M.marinum_M ----------MAIEEILDLEQLEVNIYRGSVFSPESGFLQRTFGGHVAGQ
MUL_3249|M.ulcerans_Agy99 ----------MAIEEILDLEQLEVNIYRGSVFSPESGFLQRTFGGHVAGQ
Mflv_3831|M.gilvum_PYR-GCK ----------MAIEEILDLEQIEVNIYRGGVFSPESGFLQRTFGGHVAGQ
Mvan_2565|M.vanbaalenii_PYR-1 ----------MAIEEILDLEQIEVNIYRGGVFSPESGFLQRTFGGHVAGQ
MSMEG_2938|M.smegmatis_MC2_155 ----------MAIEEILDLEQLEVNIYRGRVFSPESGFLQRTFGGHVAGQ
TH_2056|M.thermoresistible__bu ----------MAIEEILDLEQLEVNIYRGKVFSPESGFLQRTFGGHVAGQ
MAB_2891c|M.abscessus_ATCC_199 ---------MADIEEILTLEQLEKDIFRGNVTPSN---LRRTFGGQVAGQ
MLBr_01278|M.leprae_Br4923 MKRLWEKKTMSDFEELLAVLDLNRVADDLFIGSHPSKNPLRTFGGQLVAQ
:*::* : ::: : . *****::..*
Mb2637c|M.bovis_AF2122/97 SLVSAVRTVDP-RYMVHSLHGYFLRPGDAKERTVFLVERIRDGGSLCTRR
Rv2605c|M.tuberculosis_H37Rv SLVSAVRTVDP-RYMVHSLHGYFLRPGDAKERTVFLVERIRDGGSFCTRR
MAV_3483|M.avium_104 SLVSAVRTVDP-RYQVHSLHGYFLRPGDAKERTVFIVERTRDGGSFATRR
MMAR_2095|M.marinum_M SLVSAVRTVDP-RYQVHSLHGYFLRSGDAQEPTVFLVERTRDGGSFVTRR
MUL_3249|M.ulcerans_Agy99 SLVSAVRTVDP-RYQVHSLHGYFLRSGDAQEPTVFLVERTRDGGSFVTRR
Mflv_3831|M.gilvum_PYR-GCK SLVSAVRTVDP-KYQVHSLHGYFLRAGDARSPSVYTVERIRDGGSFCTRR
Mvan_2565|M.vanbaalenii_PYR-1 SLVSAVRTVDP-KFQVHSLHGYFLRAGDARSPSVYTVERIRDGGSFCTRR
MSMEG_2938|M.smegmatis_MC2_155 SLVSAVRTVAP-EFEVHSLHGYFLRPGDARAPAIYTVERIRDGGSFCTRR
TH_2056|M.thermoresistible__bu SLVSAVRTVDP-KFQVHSLHGYFLLPGDARAPTVYFVERLRDGGSFCTRR
MAB_2891c|M.abscessus_ATCC_199 SLVSAVRTVEP-EYLVHSLHGYFLRPGNPNEPTVFLVDRVRDGRSFVTRR
MLBr_01278|M.leprae_Br4923 SFVASSRTLIRDDLPPSAFSVHFINGGDTAKEIEFHVVRLRDERRFANRR
*:*:: **: :: :*: *:. : * * ** : .**
Mb2637c|M.bovis_AF2122/97 VNAVQHGETIFSMAASFQTEQEGITHQDVMPAAPPPDGLPGLNS-IKVFD
Rv2605c|M.tuberculosis_H37Rv VNAVQHGETIFSMAASFQTEQEGITHQDVMPAAPPPDGLPGLNS-IKVFD
MAV_3483|M.avium_104 VNAIQHGEIIFSMGASFQTDQEGIHHQDAMPAAPPPDGLPGLDS-IKVFD
MMAR_2095|M.marinum_M VNAVQHGEVIFSMGASFQTAQNGISHQDAMPAAPPPDDLPGLRS-VRVFD
MUL_3249|M.ulcerans_Agy99 VNAVQHGEVIFSMGASFQTAQNGISHQDAMPAAPPPDDLPGLRS-VRVFD
Mflv_3831|M.gilvum_PYR-GCK VTAIQHGETIFSMSASFQTDQSGIEHQDAMPHAVPPDDIPDFKSVSKVFD
Mvan_2565|M.vanbaalenii_PYR-1 VTAIQHGETIFSMSASFQTDQSGIEHQDAMPFAPPPDDIPDFKSSSKVFD
MSMEG_2938|M.smegmatis_MC2_155 VSAIQHGETIFSMSASFQTDQSGIEHQDAMPHAPAPEECPGFRS-GGAFD
TH_2056|M.thermoresistible__bu VSAIQHGEVIFSMSASFQTDQSGIEHQDAMPEAPPPDDLPGFRS-ARAFD
MAB_2891c|M.abscessus_ATCC_199 VTAIQDGQAIFSMSASFQTLDTGIEHQDLMPPVPDPEDLPDAQE-QDSFN
MLBr_01278|M.leprae_Br4923 VDAIQDGMLLSSAMASYMSGGCGLEHAVEPPEAAEPHTRPPIGELLRGYE
* *:*.* : * **: : *: * * . *. * . ::
Mb2637c|M.bovis_AF2122/97 DAG-----FRQFDEWDVCIVPRERLRLLPGKASQQQVWLRHRDPLPDDPV
Rv2605c|M.tuberculosis_H37Rv DAG-----FRQFDEWDVCIVPRERLRLLPGKASQQQVWLRHRDPLPDDPV
MAV_3483|M.avium_104 DAG-----FKQFEEWDVCIVPREKLQLTPGKASQQQVWFRHRDPLPDDPV
MMAR_2095|M.marinum_M DAG-----FRQFEEWDVRIVPRDLLAPLPGKASQQQVWFRHRDPLPDDPV
MUL_3249|M.ulcerans_Agy99 DAG-----FRQFEEWDVRIVPRDLLAPLPGKASQQQVWFRHRDPLPDDPV
Mflv_3831|M.gilvum_PYR-GCK DAS-----FKQFDEWDVRIVPRERLGVREGKASQQQVWFRHRDPLPDDHV
Mvan_2565|M.vanbaalenii_PYR-1 DAS-----FRQFDEWDVRIVPRDRLGVREGKASQQQVWFRHRDPMPDDHV
MSMEG_2938|M.smegmatis_MC2_155 DAG-----FAQFAEWDVRIVPRDRVQKIPGKASQQQVWFRHRDPMPDDHV
TH_2056|M.thermoresistible__bu DAG-----FAQFDEWDVRVVPRDKLKRVAGKASQQQVWFRHRDPLPDDPV
MAB_2891c|M.abscessus_ATCC_199 RDL-----FRQFAEWDIRIVPDELMDRNPRLAAQQRVWFRCRKHLPDDPV
MLBr_01278|M.leprae_Br4923 QIVQHFVNALQPVEWRYVNDPSWVMRNKGQRLAYNRVWVKALGALPNDPT
* ** * : : ::**.: :*:* .
Mb2637c|M.bovis_AF2122/97 LHICALAYMSDLTLLGSAQVNHLDVR--DQLQVASLDHAMWFMRPFRADE
Rv2605c|M.tuberculosis_H37Rv LHICALAYMSDLTLLGSAQVNHLDVR--DQLQVASLDHAMWFMRPFRADE
MAV_3483|M.avium_104 LHICALAYMSDLTLLGSAQVTHLDVR--EHLQVASLDHAMWFMRAFRADE
MMAR_2095|M.marinum_M LHICALAYMSDLTLLGSAQVTHLAER--EHLQVASLDHAMWFMRGFRADE
MUL_3249|M.ulcerans_Agy99 LHICALAYMSDLTLLGSAQVTHLAER--EHLQVASLDHAMWFMRGFRADE
Mflv_3831|M.gilvum_PYR-GCK LHICALAYMSDLTLLGSAQVNHVEER--KHLMVASLDHAMWFMRPFRADE
Mvan_2565|M.vanbaalenii_PYR-1 LHICALAYMSDLTLLGSAQVNHIEER--KHLMVASLDHAMWFMRAFRADE
MSMEG_2938|M.smegmatis_MC2_155 LHICALAYMSDLTLLGSAQVNHLDVR--KHLMVASLDHAMWFMRPFRADE
TH_2056|M.thermoresistible__bu LHICALAYMSDLTLLGSAQVNHVAER--PHLQVASLDHAMWFMRPFRADE
MAB_2891c|M.abscessus_ATCC_199 LHICALAYMSDLTLLSSSKVPHRDT----VLQTASLDHALWFLRPFRADE
MLBr_01278|M.leprae_Br4923 LHTAAMLYSTDTTVLDSVITTHGLSWGFDRIFAVSANHSVWFHRQVNFDD
** .*: * :* *:*.* . * : ..* :*::** * .. *:
Mb2637c|M.bovis_AF2122/97 WLLYDQSSPSASGGRALTRGEIFTRSGEMVAAVMQEGLTRHRRGHRSVGQ
Rv2605c|M.tuberculosis_H37Rv WLLYDQSSPSASGGRALTRGEIFTRSGEMVAAVMQEGLTRHRRGHRSVGQ
MAV_3483|M.avium_104 WLLYDQSSPSANGGRSLCQGKIFTQSGEMVAAVMQEGLTRFQRGYRQ---
MMAR_2095|M.marinum_M WLLYDQSSPSAGGGRALTHGKIFTQGGELVAAVMQEGLTRYPSGYQPGEK
MUL_3249|M.ulcerans_Agy99 WLLYDQSSPSAGGGRALTHGKIFTQGGELVAAVMQEGLTRYPSGYQPGEN
Mflv_3831|M.gilvum_PYR-GCK WLLYDQSSPSACGGRSLTQGKIYNRYGEMVAAVMQEGLTRFTRDFTPQRG
Mvan_2565|M.vanbaalenii_PYR-1 WLLYDQSSPSACGGRSLTQGKIYNRYGEMVAAVMQEGLTRYTRGYTPGHG
MSMEG_2938|M.smegmatis_MC2_155 WLLYDQTSPSACGGRALTQGRIFNQYGELVAAVMQEGLTRYKRDFTPPEE
TH_2056|M.thermoresistible__bu WLLYDQSSPSACGGRALTQGQIFNQYGEMVAAVMQEGLTRYPRNFTRTQQ
MAB_2891c|M.abscessus_ATCC_199 WMLYDETSPSAGFGRALTQGRIFNRDGTMVAAVVQEGLTRIERNPEQASV
MLBr_01278|M.leprae_Br4923 WVLYSTSSPVAADSRGLGTGHLFDRSGQLIATVVQEGVLKYFPASHR---
*:**. :** * .*.* *.:: : * ::*:*:***: :
Mb2637c|M.bovis_AF2122/97 ----------
Rv2605c|M.tuberculosis_H37Rv ----------
MAV_3483|M.avium_104 ----------
MMAR_2095|M.marinum_M ----------
MUL_3249|M.ulcerans_Agy99 ----------
Mflv_3831|M.gilvum_PYR-GCK ----------
Mvan_2565|M.vanbaalenii_PYR-1 ----------
MSMEG_2938|M.smegmatis_MC2_155 NS--------
TH_2056|M.thermoresistible__bu IQTEQTQTRP
MAB_2891c|M.abscessus_ATCC_199 ARGNMA----
MLBr_01278|M.leprae_Br4923 ----------