For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FNVTRLVDVTADDRDRVLAALRQAAATAGAHRWVVECTDPGSRNGGDILAHFRFTDETQWDQASEVAAVL RDPAITRVNGVTYHGAPVTAGTTGSVYRTLLLRVAPGTDPDAVARFEAELAAMPRHVGTILSWQLSQVTE AVGDTTWTHVFEQEFTDVAGLMGPYLMHPIHWAVVDRWFDPETTDVIIRDRVCHSFCGITGPVLS*
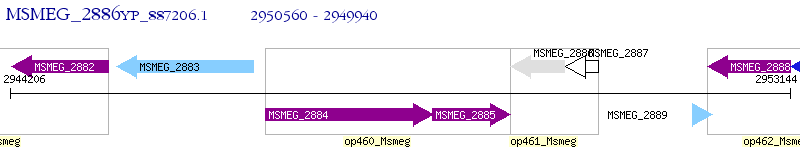
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2886 | - | - | 100% (206) | stress responsive A/B barrel domain family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4555 | - | 2e-73 | 64.42% (208) | stress responsive alpha-beta barrel domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3028 | - | 5e-76 | 66.35% (208) | stress responsive A/B barrel domain family protein |
| M. thermoresistible (build 8) | TH_0318 | - | 7e-84 | 71.50% (207) | stress responsive A/B Barrel Domain family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2885 | - | 3e-65 | 59.62% (208) | stress responsive alpha-beta barrel domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2886|M.smegmatis_MC2_155 ------MFNVTRLVDVTADDRDRVLAALRQAAATAGAHRWVVECTDPGSR
TH_0318|M.thermoresistible__bu ------MYGVTRLLDVVPEHRDRVVTALRQAATASGARRWVVERTDPRSR
Mflv_4555|M.gilvum_PYR-GCK MSTGCAMYSVIRLLDVAEPERDRVLRDVQAAAARTGAHRTVVEPTLPGSR
MAV_3028|M.avium_104 ------MYSVTRLIDIAEQDRDRMAAALRRAASDAGALHWVAGPTLPGSR
Mvan_2885|M.vanbaalenii_PYR-1 ------MYSVTRLVDVPGSDRDRVHDDLQRLAKGCGAARHLVAPTLAGSR
*:.* **:*: .***: :: * ** : :. * . **
MSMEG_2886|M.smegmatis_MC2_155 NGGDILAHFRFTDETQWD-QASEVAAVLRDPAITRVNGVTYHGAPVTAGT
TH_0318|M.thermoresistible__bu NGGDILLHLRFPDADRWSGHAPRFDELLADTAITRVNGACYQGTPVSTGQ
Mflv_4555|M.gilvum_PYR-GCK NGGDVLVHLRFQDEDEWRCHTADFEGALQDSAITRVNGAEYPGRPQ-PGH
MAV_3028|M.avium_104 NGGDLLVHLRFASKRQWDCTAACFAAILDDPAVGRVNGTTYSGTPVHAGQ
Mvan_2885|M.vanbaalenii_PYR-1 NGGDLLLHLRFDDAADWTAVAADVDAALADPAITRVNGAGYHGAPVGIAD
****:* *:** . * :. . * *.*: ****. * * * .
MSMEG_2886|M.smegmatis_MC2_155 TG--SVYRTLLLRVAPGTDPDAVARFEAELAAMPRHVGTILSWQLSQVTE
TH_0318|M.thermoresistible__bu PG--TVYRTLLLRVAPGTDAETVARFESELAAMPRYVRSIRAWQLSRVTE
Mflv_4555|M.gilvum_PYR-GCK PGPSGVYRTLLLRVLPGTEPSVVARFEDELRLMPRYVRSITAWQLSRAQD
MAV_3028|M.avium_104 KP-GTVYRTLLLAVSPDTAEDTVARFEADLRLMPRYVRTITGWQLSRVEE
Mvan_2885|M.vanbaalenii_PYR-1 RPLGGVYRTLLLSVAPDTGADVLSRFEADLQRLPGYVSTISAWQLSRPDH
******* * *.* ..::*** :* :* :* :* .****: .
MSMEG_2886|M.smegmatis_MC2_155 AVGDTTWTHVFEQEFTDVAGLMGPYLMHPIHWAVVDRWFDPETTDVIIRD
TH_0318|M.thermoresistible__bu AVGATPWTHVFEQEFTDVAGLMGPYLMHPIHWAVVDRWFDPETTEVIVRD
Mflv_4555|M.gilvum_PYR-GCK PTGDAGWTHVFEQQFTDEAGLMGPYLMHPIHWAVVDRWFDPETTDVIIRD
MAV_3028|M.avium_104 ATGASAWTHVFEQLFTDVDGLMGPYLMHPIHWAVVDRWFDPECPDVIVRD
Mvan_2885|M.vanbaalenii_PYR-1 TVGATPWTHVFEQEFSDSDGIMGPYLMHPIHWAVVDQWFDPECPNSIVRE
..* : ******* *:* *:***************:***** .: *:*:
MSMEG_2886|M.smegmatis_MC2_155 RVCHSFCGITGPVLS
TH_0318|M.thermoresistible__bu RVCHSFCVSPAPVLS
Mflv_4555|M.gilvum_PYR-GCK RVCHSFCALPAPVLD
MAV_3028|M.avium_104 RVCHSFCISTDPVLT
Mvan_2885|M.vanbaalenii_PYR-1 RVCHSFCPDPGHVLR
******* . **