For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVRPDNRLADAPMRPVGCGTCGARVLVRKASWEQTSVQWSAAARQRCVDPHRHAGQTLSLCPQLRTAIE NAVLEGAVPVLAESEDANAHRG
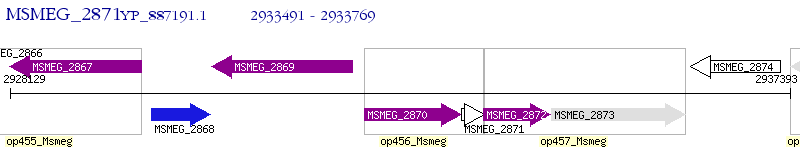
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2871 | - | - | 100% (92) | hypothetical protein MSMEG_2871 |
| M. smegmatis MC2 155 | MSMEG_5927 | - | 6e-06 | 35.48% (93) | hypothetical protein MSMEG_5927 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3557 | - | 7e-06 | 32.00% (75) | hypothetical protein Mb3557 |
| M. gilvum PYR-GCK | Mflv_4565 | - | 2e-17 | 49.40% (83) | hypothetical protein Mflv_4565 |
| M. tuberculosis H37Rv | Rv3527 | - | 8e-06 | 32.00% (75) | hypothetical protein Rv3527 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0079c | - | 1e-20 | 55.56% (81) | hypothetical protein MAB_0079c |
| M. marinum M | MMAR_5016 | - | 6e-07 | 32.89% (76) | hypothetical protein MMAR_5016 |
| M. avium 104 | MAV_3036 | - | 2e-15 | 46.51% (86) | hypothetical protein MAV_3036 |
| M. thermoresistible (build 8) | TH_0322 | - | 7e-31 | 69.05% (84) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_4090 | - | 2e-07 | 34.21% (76) | hypothetical protein MUL_4090 |
| M. vanbaalenii PYR-1 | Mvan_2870 | - | 2e-25 | 58.70% (92) | hypothetical protein Mvan_2870 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2871|M.smegmatis_MC2_155 --------------------------------------------------
TH_0322|M.thermoresistible__bu --------------------------------------------------
Mvan_2870|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0079c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4565|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3036|M.avium_104 --------------------------------------------------
Mb3557|M.bovis_AF2122/97 --MPDDQPAVPDVDRLARSMLLLHGDHHDHNDSPEQHRTCGSWSKSRDFA
Rv3527|M.tuberculosis_H37Rv --MPDDQPAVPDVDRLARSMLLLHGDHHDHNDSPEQHRTCGSWSKSRDFA
MMAR_5016|M.marinum_M MAAPDDQ-EVPDVDQLARSMLLLLGDHHDHQESSEQHHGPQPWSKSRDFT
MUL_4090|M.ulcerans_Agy99 MAAPDDQ-EVPDVDQLARSMLLLLGDHHDHQESSEQHHGRQPWSKSRDFT
MSMEG_2871|M.smegmatis_MC2_155 -----------MTVRPDNRLADAPMRPVGCGTCGARVLVRKASWEQTSVQ
TH_0322|M.thermoresistible__bu -----------VTARPDNRLLDAPMRPVTCGTCGARVLARKSSWEQTSVQ
Mvan_2870|M.vanbaalenii_PYR-1 -----------MTVRPDNRLADAPMTPVTCGACGARVLARKSSWEQTSVQ
MAB_0079c|M.abscessus_ATCC_199 ------------MIRTDNRLADSPMVPVGCERCGAQVLARKSSWQQTSVQ
Mflv_4565|M.gilvum_PYR-GCK -----------MTVRPDNRLADGPMAPVQCNRCAATVLVRKSSWHQTSVQ
MAV_3036|M.avium_104 -------------MREDVRLVDMPMAPVMCQDCGARVMARKSSWNQTSVQ
Mb3557|M.bovis_AF2122/97 DDPQRAAAVREASRAERDRYLTSGLQPVDCRFCHVTVTVKRLGPGHTAVQ
Rv3527|M.tuberculosis_H37Rv DDPQRAAAVREASRAERDRYLTSGLQPVDCRFCHVTVTVKRLGPGHTAVQ
MMAR_5016|M.marinum_M NDPQRGAAVRQASLADRERYLTAGLQPVDCRFCHVTVTVKQLGPGHTAVQ
MUL_4090|M.ulcerans_Agy99 NDPQRGAAVRQASLADRERYLTAGLQPVDCRFCHVTVTVKQLGPGHTAVQ
* : ** * * . * .:: . :*:**
MSMEG_2871|M.smegmatis_MC2_155 WSAAARQRCVDPHR---------HAGQTLSLCPQLRTAIENAVLEGAVPV
TH_0322|M.thermoresistible__bu WNAAAREMCLDQSQ---------VEGHAIPLCPALRTAIETAVVQGLLPV
Mvan_2870|M.vanbaalenii_PYR-1 WDARARSKCLSTQHLLREKCLAGDGGGVLPLCAELRVAIETAVRDGSLPV
MAB_0079c|M.abscessus_ATCC_199 WNADAESRCAQRRN----------CTGLFLVCSELRSSILDAAHSGRLPV
Mflv_4565|M.gilvum_PYR-GCK WDATATARCEERHD---ARQLAGEG-GLFLTCSALGDSVADAVVRGTLHT
MAV_3036|M.avium_104 WDADASARCIERRD---AERLAGHHRGPFLGCTALRESIQAAVTRGELAI
Mb3557|M.bovis_AF2122/97 WNTEASRRCAYFTELR----ARGGDSARTRSCPRLTDSIEHAVAEGYLEH
Rv3527|M.tuberculosis_H37Rv WNTEASRRCAYFTELR----ARGGDSARTRSCPRLTDSIEHAVAEGYLEH
MMAR_5016|M.marinum_M WNTEAAQRCAHFAEVR----RSGGDTARTKACPKMTDSIQHAIAEGVLVQ
MUL_4090|M.ulcerans_Agy99 WNTEAAQRCAHFAEVR----RSGGDTARTKACPKMTDSIQHAIAEGVVVQ
*.: * * *. : :: * * :
MSMEG_2871|M.smegmatis_MC2_155 LAES-------EDANAHRG
TH_0322|M.thermoresistible__bu LDDSAPPTTEGEPSRAHHR
Mvan_2870|M.vanbaalenii_PYR-1 LDER---------------
MAB_0079c|M.abscessus_ATCC_199 LAEEYGG------------
Mflv_4565|M.gilvum_PYR-GCK VDDA---------------
MAV_3036|M.avium_104 VDPLG--------------
Mb3557|M.bovis_AF2122/97 HDPNR--------------
Rv3527|M.tuberculosis_H37Rv HDPNR--------------
MMAR_5016|M.marinum_M HDPD---------------
MUL_4090|M.ulcerans_Agy99 HDPD---------------