For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VMAFGKRKKSAADDKRTGDQPVAAHAAPSAADEQPDEDLQGPFDIEDFDDPAVAAQGRLDLGSVLIPMPD GGQVQVELNEAGAPSAVWVVTPNGRFTIAAYAAPKTAGLWREVASELAESLRKDATSVSVQDGPWGREVV GAGNGGVVRFIGVDGYRWMVRCVVNGVPETVDALAAEARAALTDAVIRRGDTPMPVRTPLPIQLPEPMAA QLRAAAAAQAAAQQNPAAQQPQQQEGEQPQPPEPVARRSAQGSAMQQLRTITGG
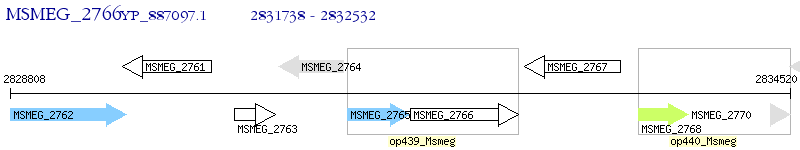
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2766 | - | - | 100% (264) | hypothetical protein MSMEG_2766 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2715c | - | 2e-92 | 63.54% (277) | hypothetical protein Mb2715c |
| M. gilvum PYR-GCK | Mflv_3932 | - | 2e-98 | 77.87% (235) | hypothetical protein Mflv_3932 |
| M. tuberculosis H37Rv | Rv2696c | - | 5e-93 | 63.90% (277) | hypothetical protein Rv2696c |
| M. leprae Br4923 | MLBr_01029 | - | 9e-88 | 63.33% (270) | hypothetical protein MLBr_01029 |
| M. abscessus ATCC 19977 | MAB_3002c | - | 4e-82 | 66.09% (230) | hypothetical protein MAB_3002c |
| M. marinum M | MMAR_2018 | - | 5e-93 | 66.42% (265) | hypothetical protein MMAR_2018 |
| M. avium 104 | MAV_3588 | - | 6e-96 | 67.54% (268) | hypothetical protein MAV_3588 |
| M. thermoresistible (build 8) | TH_1193 | - | 4e-93 | 66.67% (276) | CONSERVED HYPOTHETICAL ALANINE AND GLYCINE AND VALINE RICH |
| M. ulcerans Agy99 | MUL_3334 | - | 5e-93 | 67.17% (265) | hypothetical protein MUL_3334 |
| M. vanbaalenii PYR-1 | Mvan_2468 | - | 2e-96 | 76.25% (240) | hypothetical protein Mvan_2468 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3932|M.gilvum_PYR-GCK --------------------MGRHRSERAE----------APEEVTS---
Mvan_2468|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2766|M.smegmatis_MC2_155 -----------------MMAFGKRKKSAADDKRTGDQPVAAHAAPSA---
MMAR_2018|M.marinum_M ------------------MAFGRRTRKG----DSSGSPVEPDGPAAR---
MUL_3334|M.ulcerans_Agy99 ------------------MAFGRRTRKG----DSSGSPVEPDGPAAR---
Mb2715c|M.bovis_AF2122/97 ------------------MAFGRRTGKDGGKRKAGHAPVQPADEHVRPED
Rv2696c|M.tuberculosis_H37Rv ------------------MAFGRRTGKDGGKRKAGHAPVQPADEHVRPED
MLBr_01029|M.leprae_Br4923 MTADDDAERSDEDGAAVMATFGKRTGKD----GASRTLTEPADPEATE--
MAV_3588|M.avium_104 -----------------MAVFGRRARRG----DDEDTPVGAEIPEPDP--
TH_1193|M.thermoresistible__bu ------------------MAFGKRRRKD----KTGERVVEQQTVPAEP--
MAB_3002c|M.abscessus_ATCC_199 ------------------MALHRSGGLN-----------DLDDTADAS--
Mflv_3932|M.gilvum_PYR-GCK ---------VDTD-DEELEGPFDIDDFDDPAVAAVARLDLGSVLIPMPDA
Mvan_2468|M.vanbaalenii_PYR-1 ---------MDTH-DEELEGPFDIDDFDDPSVAALARLDLGSVLIPMPEA
MSMEG_2766|M.smegmatis_MC2_155 ---------ADEQPDEDLQGPFDIEDFDDPAVAAQGRLDLGSVLIPMPDG
MMAR_2018|M.marinum_M ------EVGEPAADDVDLVGPFDIEDFDDPSVAQLARLDLGSVLVPMPEG
MUL_3334|M.ulcerans_Agy99 ------EVGEPAADDVDLVGPFDIEDFDDPSVAQLARLDLGSVLVPMPEG
Mb2715c|M.bovis_AF2122/97 TVVASAAAASGVEDQEELQGPFDIDDFDDPSVAVLARLDLGSVLIPMPAA
Rv2696c|M.tuberculosis_H37Rv TVVASAAAASGVEDQEELQGPFDIDDFDDPSVAVLARLDLGSVLIPMPAA
MLBr_01029|M.leprae_Br4923 ---LPAASEPDSEEVDELEGPFDIDDFEDPAVAVLARLDLGSVLIPLPEG
MAV_3588|M.avium_104 ---VVAHDDADEQAADELEGPFDIDDFDDPAVAELARLDLGSVLIPMPEA
TH_1193|M.thermoresistible__bu ------DEVAGPGEDTELEGPFDIEDFDDPATAAVARLDLGSVLIPTPAT
MAB_3002c|M.abscessus_ATCC_199 --------------GETFDGPYEIEDFDSPEDAGVGRLDLGSVLIPVPET
: **::*:**:.* * .********:* *
Mflv_3932|M.gilvum_PYR-GCK GQVQVELNEAGAPSAVWVVTPNGRFTIAAYAAPKSAGLWREVASELADSL
Mvan_2468|M.vanbaalenii_PYR-1 GQVQVELNETGAPSAVWVVTPDGRFTIAAYAAPKSAGLWREVASELADSL
MSMEG_2766|M.smegmatis_MC2_155 GQVQVELNEAGAPSAVWVVTPNGRFTIAAYAAPKTAGLWREVASELAESL
MMAR_2018|M.marinum_M GQVQVELTESGVPSAVWVVTANGRFTIAAYAAPKSAGLWREVASELADSL
MUL_3334|M.ulcerans_Agy99 GQVQVELTESGVPSAVWVVTANGRFTIAAYAAPKSAGLWREVASELADSL
Mb2715c|M.bovis_AF2122/97 GQVQVELTESGVPSAVWVITPNGRYSIAAYAAPKTGGLWREVAGELADSL
Rv2696c|M.tuberculosis_H37Rv GQVQVELTESGVPSAVWVITPNGRYSIAAYAAPKTGGLWREVAGELADSL
MLBr_01029|M.leprae_Br4923 SQLQVELTDVGVPNAVWVVTANGRFTITAYAAPKTGGLWREVAGELADSL
MAV_3588|M.avium_104 GQLQVELTETGVPSAVWVVTANGRFTIAAYAAPKTGGLWREVAAELAESL
TH_1193|M.thermoresistible__bu GQVQVELNQVGAPSAVWVVTPNGRFTIAAYAAPKSAGLWREVAAELAESL
MAB_3002c|M.abscessus_ATCC_199 GQIQVELTAAGAPGAVFVLTPNGRYSVAAYAAPKSAGLWREIAGELAESL
.*:****. *.*.**:*:*.:**::::******:.*****:*.***:**
Mflv_3932|M.gilvum_PYR-GCK RKDAPKVSIEDGPWGREVVGSGGEGAA-------MVRFIGVDGYRWMIRC
Mvan_2468|M.vanbaalenii_PYR-1 RKDAPKVSIEDGPWGREVVGSAGEGAA-------VVRFIGVDGYRWMVRC
MSMEG_2766|M.smegmatis_MC2_155 RKDATSVSVQDGPWGREVVGAGNGG---------VVRFIGVDGYRWMVRC
MMAR_2018|M.marinum_M RKDSAEVSIKDGPWGREVIG-TASG---------VVRFIGVDGYRWMVRC
MUL_3334|M.ulcerans_Agy99 RKDSAEVSIKDGLWGREVIG-TASG---------VVRFIGVDGYRWMVRC
Mb2715c|M.bovis_AF2122/97 RKDSAKVSIKDGPWGREVIG-IAAG---------VVRFIGVNGYRWMIRC
Rv2696c|M.tuberculosis_H37Rv RKDSAKVSIKDGPWGREVIG-IAAG---------VVRFIGVDGYRWMIRC
MLBr_01029|M.leprae_Br4923 RNDSAKVTVKDGPWGREVVG-TNTG---------VVRFIGVDGYRWMIRC
MAV_3588|M.avium_104 RNDSAKVSIQDGPWGREVVG-TAAG---------VVRFIGVDGYRWMIRC
TH_1193|M.thermoresistible__bu RKDATDVRIEDGPWGREVVG-TAPGQANESGQPTLVRFIGVDGYRWMIRC
MAB_3002c|M.abscessus_ATCC_199 RKEAAEVSIADGPWGREVVGAAQGG---------SVRFIGVDGYRWMIRC
*:::..* : ** *****:* * ******:*****:**
Mflv_3932|M.gilvum_PYR-GCK VVNGPFERIAELTEQARDALADTVVRRGDTPLPVRTPLPVTLPEPMAAQL
Mvan_2468|M.vanbaalenii_PYR-1 VVNGPYERMAELAEQARNALADTVVRRGDTPLPVRTPLPVQLPEPMAAQL
MSMEG_2766|M.smegmatis_MC2_155 VVNGVPETVDALAAEARAALTDAVIRRGDTPMPVRTPLPIQLPEPMAAQL
MMAR_2018|M.marinum_M VVNGPHETIDTLTDEAREALADTVIRRGDTPLPVRTPLPVQLPEPMVEQL
MUL_3334|M.ulcerans_Agy99 VVNGPHETIDTLTDEAREALADTVIRRGDTPLPVRTPLPVQLPEPMVEQL
Mb2715c|M.bovis_AF2122/97 VVNGPQETVDALTEEAREALADTVVRRGDTPLPVRTPLPVHLPEPMAAQL
Rv2696c|M.tuberculosis_H37Rv VVNGPQETVDALTEEAREALADTVVRRGDTPLPVRTPLPVHLPEPMAAQL
MLBr_01029|M.leprae_Br4923 VVNGPLETIDVLSEEARAALADTVVRRGDTPLPVRTPLPVQLPEQMAEQL
MAV_3588|M.avium_104 VVNGTHETMEALEQEARAALADTVVRRGDTPLPVRTPLPVQLPEPMAQQL
TH_1193|M.thermoresistible__bu VVNGAADKMHALAEEARDALADTVVRRGDTPLPVRTPLPVELPEPMAAQL
MAB_3002c|M.abscessus_ATCC_199 VLQGPAETFDALADEAREAVADTVVRRDDSPYPVRTPLPVVLPEPMLEQL
*::* : . * :** *::*:*:**.*:* *******: *** * **
Mflv_3932|M.gilvum_PYR-GCK -AAAQQQAAAQQQQAATEQQPPAQQQPP-----AQQEPPQPNARRSAPGS
Mvan_2468|M.vanbaalenii_PYR-1 -AAAQQQAVAQQQAAAQQQQAPAQQQAPEQPAPAQQEPPQPNARRSTMGS
MSMEG_2766|M.smegmatis_MC2_155 RAAAAAQAAAQQNPAAQQ---PQQQEGE------QPQPPEPVARRSAQGS
MMAR_2018|M.marinum_M RQAAAAQHEQGQQ-----------------------EPDQPAPRRGAEGS
MUL_3334|M.ulcerans_Agy99 RQAAATQHEQGQQG---Q-----------------QEPDQPVPRRGAEGS
Mb2715c|M.bovis_AF2122/97 REAAAAQADTQRQ------------------------AAAGVARRGAQGS
Rv2696c|M.tuberculosis_H37Rv REAAAAQADTQRQ------------------------AAAGVARRGAQGS
MLBr_01029|M.leprae_Br4923 REAAVAQQSAQHADARQQ-----------------S--RELAPRRGAAGS
MAV_3588|M.avium_104 REAAAAQQAAAQQAQSQQ-----------------SPGNEPAARRSADGS
TH_1193|M.thermoresistible__bu RAAAAQQAQQSAPAQQQT--------------------QQPGARR-TQGS
MAB_3002c|M.abscessus_ATCC_199 QQAQAQAMAQAEAEAPAQEPAP-------------QDDPTPAARRSVAGS
* .** . **
Mflv_3932|M.gilvum_PYR-GCK AMQQLR-TITGG
Mvan_2468|M.vanbaalenii_PYR-1 AMQQLR-TITGG
MSMEG_2766|M.smegmatis_MC2_155 AMQQLR-TITGG
MMAR_2018|M.marinum_M AMQQLR-STTGG
MUL_3334|M.ulcerans_Agy99 AMQQLR-STTGG
Mb2715c|M.bovis_AF2122/97 AMQQLR-STTGG
Rv2696c|M.tuberculosis_H37Rv AMQQLR-STTGG
MLBr_01029|M.leprae_Br4923 AMQQLH-NTTGG
MAV_3588|M.avium_104 AMQQLR-TTTGG
TH_1193|M.thermoresistible__bu AMQQLRSTITGG
MAB_3002c|M.abscessus_ATCC_199 AMQQLRSTITGG
*****: . ***