For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSTSLAVVRLDRELPMPTRAHDGDAGVDLYSAENVELAPGQRALVSTGIAVAIPHGMVGLVHPRSGLAAR VGLSIVNSPGTIDAGYRGEIKVSLINLDPQTPVVISRGDRIAQLLVQRVELPELVEVTSFDEAGLADTTR GDGGHGSSGGHASL
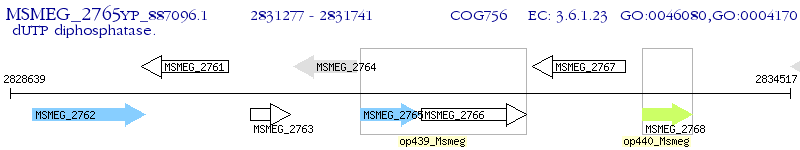
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2765 | dut | - | 100% (154) | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| M. smegmatis MC2 155 | MSMEG_0678 | dcd | 2e-05 | 28.00% (125) | deoxycytidine triphosphate deaminase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2716c | dut | 5e-72 | 85.06% (154) | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| M. gilvum PYR-GCK | Mflv_3933 | dut | 2e-74 | 87.01% (154) | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| M. tuberculosis H37Rv | Rv2697c | dut | 5e-72 | 85.06% (154) | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| M. leprae Br4923 | MLBr_01028 | dut | 7e-72 | 85.06% (154) | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| M. abscessus ATCC 19977 | MAB_3003c | dut | 2e-67 | 83.33% (150) | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| M. marinum M | MMAR_2017 | dut | 3e-71 | 85.71% (154) | deoxyuridine 5'-triphosphate nucleotidohydrolase DutT |
| M. avium 104 | MAV_3589 | dut | 5e-71 | 85.06% (154) | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| M. thermoresistible (build 8) | TH_1192 | dut | 1e-71 | 84.42% (154) | PROBABLE DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE DUT |
| M. ulcerans Agy99 | MUL_3335 | dut | 4e-71 | 85.71% (154) | deoxyuridine 5'-triphosphate nucleotidohydrolase |
| M. vanbaalenii PYR-1 | Mvan_2467 | dut | 6e-77 | 90.26% (154) | deoxyuridine 5'-triphosphate nucleotidohydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2716c|M.bovis_AF2122/97 MSTTLAIVRLDPGLPLPSRAHDGDAGVDLYSAEDVELAPGRRALVRTGVA
Rv2697c|M.tuberculosis_H37Rv MSTTLAIVRLDPGLPLPSRAHDGDAGVDLYSAEDVELAPGRRALVRTGVA
MAV_3589|M.avium_104 MSTSLAIVRLDPGLPLPSRAHEGDAGVDLYSAEDVRLEPGRRALVRTGVA
MMAR_2017|M.marinum_M MSNSLAVVRLDPGLPLPSRAHDGDAGVDLYSAEDVVLPPGQRALVRTGVA
MUL_3335|M.ulcerans_Agy99 MSNSLAVVRLDPGLPLPSRAHDGDAGVDLYSAEDVVLPPGQRALVRTGVA
MLBr_01028|M.leprae_Br4923 MSTSLAVVRLDPGLPLPSRAHDGDAGVDLYSVEDVKLAPGQRALVRTGLA
Mflv_3933|M.gilvum_PYR-GCK MSTSLAVVRLDPDLPLPSRAHDGDAGVDLYSAQDVELGPGERALVPTGVA
Mvan_2467|M.vanbaalenii_PYR-1 MPTTLAVVRLDRDLPLPSRAHDGDAGVDLYSAQDVELAPGQRALVPTGIA
TH_1192|M.thermoresistible__bu VSTSLPIVRLDRDLPMPTRAHHGDAGVDLFSAVDVELAPQQRALVPTGIA
MSMEG_2765|M.smegmatis_MC2_155 MSTSLAVVRLDRELPMPTRAHDGDAGVDLYSAENVELAPGQRALVSTGIA
MAB_3003c|M.abscessus_ATCC_199 ----MAIVRLDRELPLPSRAHADDAGVDLYSAEDVVIEPGRRTLVGTGIA
:.:**** **:*:*** .******:*. :* : * .*:** **:*
Mb2716c|M.bovis_AF2122/97 VAVPFGMVGLVHPRSGLATRVGLSIVNSPGTIDAGYRGEIKVALINLDPA
Rv2697c|M.tuberculosis_H37Rv VAVPFGMVGLVHPRSGLATRVGLSIVNSPGTIDAGYRGEIKVALINLDPA
MAV_3589|M.avium_104 VAIPFGMVGLVHPRSGLAARVGLSIVNSPGTIDAGYRGEIKVALINLDPA
MMAR_2017|M.marinum_M VAIPFGMVGLVHPRSGLASRVGLSIVNSPGTIDAGYRGELKVALINLDPA
MUL_3335|M.ulcerans_Agy99 VAIPFGMVGLVHPRSGLASRVGLSIVNSPGTIDAGYRGELKVVLINLDPA
MLBr_01028|M.leprae_Br4923 VAIPFGMVGLIHPRSGLAVRVGLSIVNSPGTVDAGYRGEIKVALINLDPV
Mflv_3933|M.gilvum_PYR-GCK VAIPHGMVGLIHPRSGLAARVGLSIVNTPGTVDAGYRGEIKVCLINLDTS
Mvan_2467|M.vanbaalenii_PYR-1 VAIPHGMVGLIHPRSGLAARVGLSIVNSPGTVDAGYRGEIKVSLINLDPA
TH_1192|M.thermoresistible__bu VAIPVGMVGLIHPRSGLAARLGLSIVNAPGTVDAGYRGEIKVALINLDPE
MSMEG_2765|M.smegmatis_MC2_155 VAIPHGMVGLVHPRSGLAARVGLSIVNSPGTIDAGYRGEIKVSLINLDPQ
MAB_3003c|M.abscessus_ATCC_199 VAIPSGMVGLVHPRSGLAARVGLSIVNSPGTIDAGYRGEVKVNLINLDSE
**:* *****:******* *:******:***:*******:** *****.
Mb2716c|M.bovis_AF2122/97 APIVVHRGDRIAQLLVQRVELVELVEVSSFDEAGLASTSRGDGGHGSSGG
Rv2697c|M.tuberculosis_H37Rv APIVVHRGDRIAQLLVQRVELVELVEVSSFDEAGLASTSRGDGGHGSSGG
MAV_3589|M.avium_104 EPIVVHRGDRIAQLLVQRVELVELVEVSSFDEAGLAGTSRGDGGHGSSGG
MMAR_2017|M.marinum_M TPIVVNRGDRIAQLLVQRVELLELVEVSSFDEAGLAATSRGDGGHGSSGG
MUL_3335|M.ulcerans_Agy99 TPIVVNRGDRIAQLLVQRVELLELVEVSSFDEAGLAATSRGDGGHGSSGG
MLBr_01028|M.leprae_Br4923 EPLVVHRGDRIAQLLVQRVELVELVEVSSFDEAGLAETSRGDGGHGSSGG
Mflv_3933|M.gilvum_PYR-GCK TPITIGRGDRIAQLLVQRVELPELVEVTSFDEAGLADTTRGDGGYGSSGG
Mvan_2467|M.vanbaalenii_PYR-1 APIAIRRGDRIAQLLVQRVELPELVEVTSFDEAGLADTTRGDGGHGSSGG
TH_1192|M.thermoresistible__bu TPISVRRGDRIAQLLVQRVELPELVEVASFDEVGLADTSRGDGGYGSSGG
MSMEG_2765|M.smegmatis_MC2_155 TPVVISRGDRIAQLLVQRVELPELVEVTSFDEAGLADTTRGDGGHGSSGG
MAB_3003c|M.abscessus_ATCC_199 VPIVIARGDRIAQLLVQQVELPELVEVDSFDEAGLAVTTRGAGGYGSSGG
*: : ***********:*** ***** ****.*** *:** **:*****
Mb2716c|M.bovis_AF2122/97 HASL
Rv2697c|M.tuberculosis_H37Rv HASL
MAV_3589|M.avium_104 HASL
MMAR_2017|M.marinum_M HASL
MUL_3335|M.ulcerans_Agy99 HASL
MLBr_01028|M.leprae_Br4923 HASL
Mflv_3933|M.gilvum_PYR-GCK HASL
Mvan_2467|M.vanbaalenii_PYR-1 HASL
TH_1192|M.thermoresistible__bu HASL
MSMEG_2765|M.smegmatis_MC2_155 HASL
MAB_3003c|M.abscessus_ATCC_199 HASL
****