For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADNENPQQRYQPDQSGMYELEFPAPQLSSSDGRGPVLIHALEGFSDAGHAIRLAAEHLKKSLDTELVASF AIDELLDYRSRRPLMTFKTDHFTAYEEPELNLYALHDTVGTPFLLLAGLEPDLRWERFITAVRLLAEQLG VRQVIGLGTIPMAVPHTRPVNLTAHSNNKELIAEHTPWVGEVQVPASVSNLLEFRMAQHGHEVVGFTVHV PHYLAQTAYPPAAEALLAEVARTGSLELPLAALSEAGAEVYTKINEQVEASPEVAQVVSALERQYDAFIA AQENRSLLARDEDLPSGDELGAEFERFLAQQAGEKFKDDKYKDGHGEGFGGRIRGTVTTTDDVPDQG*
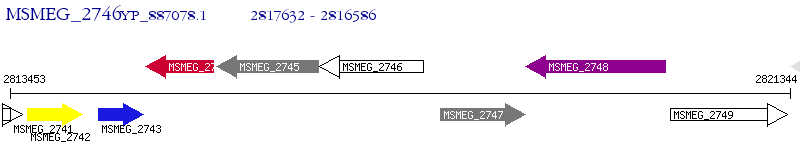
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2746 | - | - | 100% (348) | hypothetical protein MSMEG_2746 |
| M. smegmatis MC2 155 | MSMEG_4186 | - | 7e-27 | 28.11% (281) | hypothetical protein MSMEG_4186 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2733 | - | 1e-148 | 81.73% (312) | hypothetical protein Mb2733 |
| M. gilvum PYR-GCK | Mflv_3951 | - | 1e-153 | 82.82% (326) | hypothetical protein Mflv_3951 |
| M. tuberculosis H37Rv | Rv2714 | - | 1e-146 | 81.09% (312) | hypothetical protein Rv2714 |
| M. leprae Br4923 | MLBr_01009 | - | 1e-147 | 80.83% (313) | hypothetical protein MLBr_01009 |
| M. abscessus ATCC 19977 | MAB_3033 | - | 1e-139 | 74.77% (321) | hypothetical protein MAB_3033 |
| M. marinum M | MMAR_1998 | - | 1e-151 | 81.37% (322) | hypothetical protein MMAR_1998 |
| M. avium 104 | MAV_3608 | - | 1e-146 | 78.75% (320) | hypothetical protein MAV_3608 |
| M. thermoresistible (build 8) | TH_0184 | - | 1e-151 | 84.66% (313) | CONSERVED HYPOTHETICAL ALANINE AND LEUCINE RICH PROTEIN |
| M. ulcerans Agy99 | MUL_3357 | - | 1e-150 | 81.06% (322) | hypothetical protein MUL_3357 |
| M. vanbaalenii PYR-1 | Mvan_2446 | - | 1e-152 | 82.52% (326) | hypothetical protein Mvan_2446 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2746|M.smegmatis_MC2_155 ----MADNENPQQRYQPDQSGMYELEFPAPQLSSSDGRGPVLIHALEGFS
TH_0184|M.thermoresistible__bu ---------------------MYELEFPAPKLSSEDGKGPVLVHALEGFS
Mflv_3951|M.gilvum_PYR-GCK ---MADDAHDSGSRYQPEQTGMYELEFPGPQLSTPDGRGPVLIHALEGFS
Mvan_2446|M.vanbaalenii_PYR-1 ---MADDVHDPGHEYQPEQTGMYELEFPGPQLSSPDGRGPVLIHALEGFS
MMAR_1998|M.marinum_M --MAHDQDPGEAQDYQPGQSGMYELEFPAPQLSTSDGRGPVLVHALEGFS
MUL_3357|M.ulcerans_Agy99 --MAHDQDPGEAQDYQPGQSGMYELEFPAPQLSTSDGRGPVLVHALEGFS
Mb2733|M.bovis_AF2122/97 --MARDQGADEAREYEPGQPGMYELEFPAPQLSSSDGRGPVLVHALEGFS
Rv2714|M.tuberculosis_H37Rv --MARDQGADEAREYEPGQPGMYELEFPAPQLSSSDGRGPVLVHALEGFS
MLBr_01009|M.leprae_Br4923 MSHSHYQDPDDEQHYQPGQPGMYVLEFPAPQLLASDGRGPVLIHALEGFS
MAV_3608|M.avium_104 --MTHRQDAGD--QYQPGQAGMYELELPAPQLSTSDGRGPVLVHALEGFS
MAB_3033|M.abscessus_ATCC_1997 ---MTSNEGVEPQPYKPDQSGMYELEFPGPQLASADGRGPVLVHALQGFS
** **:*.*:* : **:****:***:***
MSMEG_2746|M.smegmatis_MC2_155 DAGHAIRLAAEHLKKSLDTELVASFAIDELLDYRSRRPLMTFKTDHFTAY
TH_0184|M.thermoresistible__bu DAGHAIRLAAEHLKNTLTTELVASFAIDELLDYRSRRPLMTFKTDHFTRY
Mflv_3951|M.gilvum_PYR-GCK DAGHAIKLAAAHLKNSLDTELVASFAIDELLDYRSRRPLMTFKTDHFTHY
Mvan_2446|M.vanbaalenii_PYR-1 DAGHAIRLSAQHLKDTLDTELVASFAIDELLDYRSRRPLMTFKTDHFTHY
MMAR_1998|M.marinum_M DAGHAIRLAATHLKDGLDTELVASFAIDELLDYRSRRPMMTFKTDHFTKY
MUL_3357|M.ulcerans_Agy99 DAGHAIRLAATHLKDGLDTELVASFAIDELLDYRSRRSMMTFKTDHFTKY
Mb2733|M.bovis_AF2122/97 DAGHAIRLAAAHLKAALDTELVASFAIDELLDYRSRRPLMTFKTDHFTHS
Rv2714|M.tuberculosis_H37Rv DAGHAIRLAAAHLKAALDTELVASFAIDELLDYRSRRPLMTFKTDHFTHS
MLBr_01009|M.leprae_Br4923 DAGHAIRLAATHLKAALNTELVASFAIDELLDYRSRRPLMTFKTDHFTHY
MAV_3608|M.avium_104 DAGHAIRLAAKHLKAALDSELVASFAIDELLDYRSRRPLMTFKTDHFTHY
MAB_3033|M.abscessus_ATCC_1997 DSGHAVKLAAAHLRQTLESELVASFAIDDLLDYRSRRPVMTFKSDHFTEY
*:***::*:* **: * :*********:********.:****:****
MSMEG_2746|M.smegmatis_MC2_155 EEPELNLYALHDTVGTPFLLLAGLEPDLRWERFITAVRLLAEQLGVRQVI
TH_0184|M.thermoresistible__bu EEPELNLYALHDTAGTPFLLLTGLEPDLKWERFVSAVRLLAERLGVRRVI
Mflv_3951|M.gilvum_PYR-GCK DEPELNLYALHDSVGTPFLLLSGMEPDLRWERFVTAIRLLAERLGVRRVI
Mvan_2446|M.vanbaalenii_PYR-1 DQPELNLYALRDTAGTPFLLLAGLEPDLRWERFITAVRLLSERLGVRRVI
MMAR_1998|M.marinum_M DDPELSLYALRDSVGTPFLLLAGMEPDLKWERFITAVRLLAERLGVRQTI
MUL_3357|M.ulcerans_Agy99 DDPELSLYALRDSVGTPFLLLAGMEPDLKWERFITAVRLLAERLGVRQTI
Mb2733|M.bovis_AF2122/97 DDPELSLYALRDSIGTPFLLLAGLEPDLKWERFITAVRLLAERLGVRQTI
Rv2714|M.tuberculosis_H37Rv DDPELSLYALRDSIGTPFLLLAGLEPDLKWERFITAVRLLAERLGVRQTI
MLBr_01009|M.leprae_Br4923 DDPELSLYALRDSVGTPFLLLAGMEPDLKWERFITAVRLLAERLGVRQTI
MAV_3608|M.avium_104 DDPELSLYALRDSVGTPFLLLAGLEPDLKWERFITAVRLLAERLGVRQTI
MAB_3033|M.abscessus_ATCC_1997 ATPELNLYALKDTKGTPFLLLAGLEPDLKWERFVNAIRLLAEQLGVRKTI
***.****:*: *******:*:****:****:.*:***:*:****:.*
MSMEG_2746|M.smegmatis_MC2_155 GLGTIPMAVPHTRPVNLTAHSNNKELIAEHTPWVGEVQVPASVSNLLEFR
TH_0184|M.thermoresistible__bu GLGTIPMAVPHTRPVTLTAHSNNKELIAEHTAWVGEVQVPGSVSNLLEFR
Mflv_3951|M.gilvum_PYR-GCK GLGSIPMAVPHTRPMTLTAHSNDKELIAEHQPWVGEVQVPGSASNLLEFR
Mvan_2446|M.vanbaalenii_PYR-1 GLGSIPMAVPHTRPMTLTAHSNDKELIAEHQPWVNEVQVPGSASNLLEFR
MMAR_1998|M.marinum_M GLGTVPMAVPHTRPVTMTAHSNNPELIADFQPWISEIQVPASASNLLEYR
MUL_3357|M.ulcerans_Agy99 GLGTVPMAVPHTRPVTMTAHSNNPELIADFQPWISEIQVPASASNLLEYR
Mb2733|M.bovis_AF2122/97 GLGTVPMAVPHTRPITMTAHSNNRELISDFQPWISEIQVPGSASNLLEYR
Rv2714|M.tuberculosis_H37Rv GLGTVPMAVPHTRPITMTAHSNNRELISDFQPSISEIQVPGSASNLLEYR
MLBr_01009|M.leprae_Br4923 SLGTVPMAVPHTRPITLTAHSNNGELIADFTPWITEIQVPGSASNLLEYR
MAV_3608|M.avium_104 GLGTVPMAVPHTRPITMTAHSNNPELIKNFQPWIAEIQVPGSASNLLEYR
MAB_3033|M.abscessus_ATCC_1997 GLGAIPMAVPHTRPITLTAHGNDRKTLDEHPGWIDEVQVPGSASNLLEFR
.**::*********:.:***.*: : : :. : *:***.*.*****:*
MSMEG_2746|M.smegmatis_MC2_155 MAQHGHEVVGFTVHVPHYLAQTAYPPAAEALLAEVARTGSLELPLAALSE
TH_0184|M.thermoresistible__bu MGQHGFEVVGYTVHVPHYLAQTAYPPAAEALLAEVAKAGSLELPLAELAK
Mflv_3951|M.gilvum_PYR-GCK MAQHGYEVVGFTVHVPHYLAQTDYPSAAETLLAEVARTASLDIPTAELTT
Mvan_2446|M.vanbaalenii_PYR-1 MAQHGYEVVGFTVHVPHYLAQTDYPSAAETLLSEVARNGSLEIPTTKLTQ
MMAR_1998|M.marinum_M MAQHGHEVVGFTVHVPHYLTQTDYPAAAQALLEQVAKTGSLDLPLAALTD
MUL_3357|M.ulcerans_Agy99 MAQHGHEVVGFTVHVPHYLTQTDYPAAAQALLEQVAKTGSLDLPLAALTD
Mb2733|M.bovis_AF2122/97 MAQHGHEVVGFTVHVPHYLTQTDYPAAAQALLEQVAKTGSLQLPLAALAE
Rv2714|M.tuberculosis_H37Rv MAQHGHEVVGFTVHVPHYLTQTDYPAAAQALLEQVAKTGSLQLPLAVLAE
MLBr_01009|M.leprae_Br4923 MGQHGHEVVGFTVHVPHYLTQTDYPAAAQALLEQVAKTGALQLPLSALAE
MAV_3608|M.avium_104 MAQHGHEVVGYTVHVPHYLTQTDYPAAAQALLEQVAKTASLELPLTALSE
MAB_3033|M.abscessus_ATCC_1997 LAQHGHDVVGFAVHVPHYLAQTDYPEASQRLLEEVARTGDLDLPLQELSE
:.***.:***::*******:** ** *:: ** :**: . *::* *:
MSMEG_2746|M.smegmatis_MC2_155 AGAEVYTKINEQVEASPEVAQVVSALERQYDAFIAAQENRSLLARDEDLP
TH_0184|M.thermoresistible__bu AGAEVHDRINEQVEASPEVAQVVAALERQYDAFITAQENRSLLAHDEDLP
Mflv_3951|M.gilvum_PYR-GCK AAAVVFDKINEQVTASAEVAQVVDALERQYDAFVAAQENRSLLARDEDLP
Mvan_2446|M.vanbaalenii_PYR-1 AAAEVFDKINEQVAGSAEVAQVVEALERQYDAFVAAQENRSLLARDEDLP
MMAR_1998|M.marinum_M ASAQVGAKIDEQVQASAEVAQVVAALERQYDAFIDAQENRSLLTRDEDLP
MUL_3357|M.ulcerans_Agy99 ASAQVGAKIDEQVQASAEVAQVVAALERQYDAFIDAQENRSLLTRDEDLP
Mb2733|M.bovis_AF2122/97 AAAEVQAKIDEQVQASAEVAQVVAALERQYDAFIDAQENRSLLTRDEDLP
Rv2714|M.tuberculosis_H37Rv AAAEVQAKIDEQVQASAEVAQVVAALERQYDAFIDAQENRSLLTRDEDLP
MLBr_01009|M.leprae_Br4923 AAAEIRAKIDEQVQASTEVAQVVAALERQYDAFIDAQENRSLLRRDEDLP
MAV_3608|M.avium_104 AAEVIRAKIDEQVEASAEVAQVVASLERQYDAFIDAQENRSLLTRDGDLP
MAB_3033|M.abscessus_ATCC_1997 AAAKVRNQINEQVEGSEEVAQVVQALERQYDAFVAAQENRSLLARDEELP
*. : :*:*** .* ****** :********: ******** :* :**
MSMEG_2746|M.smegmatis_MC2_155 SGDELGAEFERFLAQQAGEKFKDDKYKDGHGEGFGGRIRGTVTTTDDVPD
TH_0184|M.thermoresistible__bu SGEELAGEFEKFLAQQNRESNPDGFRDDGDTPGF----------------
Mflv_3951|M.gilvum_PYR-GCK SGEELGAEFERFLAQQAGEKKRKD-GDDQPREGL----------------
Mvan_2446|M.vanbaalenii_PYR-1 SGEELGAEFERFLAQQAGEKKRKD-GDD-PREGL----------------
MMAR_1998|M.marinum_M SGDELGAEFERFLAQQAEKK-----FDDDDQP------------------
MUL_3357|M.ulcerans_Agy99 SGDELGAEFERFLAQQAEKK-----FDDDDQP------------------
Mb2733|M.bovis_AF2122/97 SGDELGAEFERFLAQQAEKK-----SDDDPT-------------------
Rv2714|M.tuberculosis_H37Rv SGDELGAEFERFLAQQAEKK-----SDDDPT-------------------
MLBr_01009|M.leprae_Br4923 SGDELGAEFERFLAQQAEKK-----RDDDLT-------------------
MAV_3608|M.avium_104 SGDELGAEFERFLAQQAEKK-----FDDDDDQA-----------------
MAB_3033|M.abscessus_ATCC_1997 SGDELAGEFERFLAEQAKFG------DDPSGPSPEGPIL-----------
**:**..***:***:* .*
MSMEG_2746|M.smegmatis_MC2_155 QG
TH_0184|M.thermoresistible__bu --
Mflv_3951|M.gilvum_PYR-GCK --
Mvan_2446|M.vanbaalenii_PYR-1 --
MMAR_1998|M.marinum_M --
MUL_3357|M.ulcerans_Agy99 --
Mb2733|M.bovis_AF2122/97 --
Rv2714|M.tuberculosis_H37Rv --
MLBr_01009|M.leprae_Br4923 --
MAV_3608|M.avium_104 --
MAB_3033|M.abscessus_ATCC_1997 --