For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VIFAKGHGTENDFVVLPDLDAALSLTPSAVAALCDRRRGLGADGVLRVTRAGAARAAGVFDRLPDGVSAG DWYMDYRNADGSIAQMCGNGVRVFAHYLRASGLESADEFVVGSLAGPRPVVLHGFDPARATTAEVTVEMG KANQFGAGSAVVGGRSFDGLAVDVGNPHLACVGITADDLAALDVAAPVSFDPALFPDGVNVEVLTASVDG AVSMRVHERGVGETRSCGTGTVAATVAALAHDGADTGTLRVRIPGGEVTVTVTESTSYLRGPSVLVARGE LDPHWWHAVAG
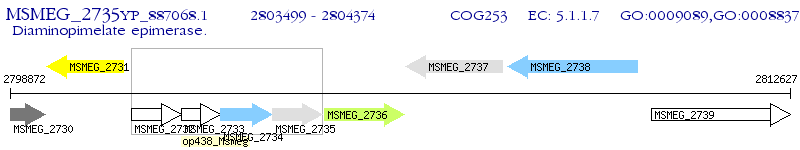
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2735 | dapF | - | 100% (291) | diaminopimelate epimerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2745c | dapF | 1e-123 | 76.29% (291) | diaminopimelate epimerase |
| M. gilvum PYR-GCK | Mflv_3961 | dapF | 1e-121 | 76.55% (290) | diaminopimelate epimerase |
| M. tuberculosis H37Rv | Rv2726c | dapF | 1e-123 | 76.29% (291) | diaminopimelate epimerase |
| M. leprae Br4923 | MLBr_00996 | dapF | 1e-117 | 70.61% (296) | diaminopimelate epimerase |
| M. abscessus ATCC 19977 | MAB_3043c | dapF | 1e-104 | 65.31% (294) | diaminopimelate epimerase |
| M. marinum M | MMAR_1987 | dapF | 1e-121 | 75.69% (288) | diaminopimelate epimerase DapF |
| M. avium 104 | MAV_3619 | dapF | 1e-121 | 75.34% (292) | diaminopimelate epimerase |
| M. thermoresistible (build 8) | TH_1519 | dapF | 1e-132 | 82.41% (290) | PROBABLE DIAMINOPIMELATE EPIMERASE DAPF (DAP EPIMERASE) |
| M. ulcerans Agy99 | MUL_3369 | dapF | 1e-121 | 75.69% (288) | diaminopimelate epimerase |
| M. vanbaalenii PYR-1 | Mvan_2435 | dapF | 1e-128 | 79.31% (290) | diaminopimelate epimerase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2735|M.smegmatis_MC2_155 MIFAKGHGTENDFVVLPDLDAALSLTPSAVAALCDRRRGLGADGVLRVTR
TH_1519|M.thermoresistible__bu VRFAKGHGTENDFVVLPDPDARLTLTPAGVAALCDRRRGLGADGVLRVTT
Mb2745c|M.bovis_AF2122/97 MIFAKGHGTQNDFVLLPDVDAELVLTAARVAALCDRRKGLGADGVLRVTT
Rv2726c|M.tuberculosis_H37Rv MIFAKGHGTQNDFVLLPDVDAELVLTAARVAALCDRRKGLGADGVLRVTT
MMAR_1987|M.marinum_M MIFAKGHGTQNDFVLLPDVAARLSLTAAQVAALCDRRRGLGADGILRVTT
MUL_3369|M.ulcerans_Agy99 MIFAKGHGTQNDFVLLPDVAARLSLTAAQVAALCDRRRGLGADGILRVTT
MAV_3619|M.avium_104 MKFAKGHGTENDFVLLCDTPAELRLTAAGVAALCDRRRGLGADGVLRVTT
MLBr_00996|M.leprae_Br4923 MIFAKGHGTQNDFVVLPDVEADVTFTAAQVAALCNRRQGLGADGVLRVTT
Mvan_2435|M.vanbaalenii_PYR-1 MKFAKGHGTQNDFVLLPDLTADLALTPAAVAALCDRRQGLGADGVLRVTT
Mflv_3961|M.gilvum_PYR-GCK MKFAKGHGTQNDFVLLPDLNAQYSLTARAVAALCDRRRGLGADGLLRVTT
MAB_3043c|M.abscessus_ATCC_199 MKFTKGHGTQNDFVVLPDVHIKRDLSVAAVQALCDRQRGLGADGVLRVTT
: *:*****:****:* * :: * ***:*::******:****
MSMEG_2735|M.smegmatis_MC2_155 AGAARAAGVFDRLPDGVSAGDWYMDYRNADGSIAQMCGNGVRVFAHYLRA
TH_1519|M.thermoresistible__bu AGAARAAGVLDRLPDGVAADDWYMDYRNADGSIAQMCGNGVRVFAHYLRA
Mb2745c|M.bovis_AF2122/97 AGAAQAVGVLDSLPEGVRVTDWYMDYRNADGSAAQMCGNGVRVFAHYLRA
Rv2726c|M.tuberculosis_H37Rv AGAAQAVGVLDSLPEGVRVTDWYMDYRNADGSAAQMCGNGVRVFAHYLRA
MMAR_1987|M.marinum_M ADAAVAAGVLHRLPDGVSPGDWYMDYRNADGSTAQMCGNGVRVFAHYLRA
MUL_3369|M.ulcerans_Agy99 ADAAVAAGVLHRLPDGVSPGDWYMDYRNADGSTAQMCGNGVRVFAHYLRA
MAV_3619|M.avium_104 AGAAAAAGVLDRLPDGVAGDDWYMDYRNSDGSVAQMCGNGVRVFAHYLRA
MLBr_00996|M.leprae_Br4923 AGAAVTAGVLEHLPDGVSCSDWYMDYRNADGSVAQMCGNGVRVFAHYLRA
Mvan_2435|M.vanbaalenii_PYR-1 AGAARAAGVFDRLPEGVVEGDWYMDYRNADGSIAQMCGNGVRVFAHYLRA
Mflv_3961|M.gilvum_PYR-GCK AGAALAAGVFDRLPEGVAEDDWFMDYRNADGSIAEMCGNGVRVFAHYLHA
MAB_3043c|M.abscessus_ATCC_199 VGAALEGGVLAEKPAGVSSEDWFMDYRNADGSIAEMCGNGVRVFAHYLRS
..** **: * ** **:*****:*** *:*************::
MSMEG_2735|M.smegmatis_MC2_155 SGLESADEFVVGSLAGPRPVVLHGFDPARATTAEVTVEMGKANQFGAG--
TH_1519|M.thermoresistible__bu SGLESRDEFVVGSPAGPRPVVLHHSD---ATTADVTVEMGKANQSGTG--
Mb2745c|M.bovis_AF2122/97 SGLEVRDEFVVGSLAGPRPVTCHHVE---AAYADVSVDMGKANRLGAG--
Rv2726c|M.tuberculosis_H37Rv SGLEVRDEFVVGSLAGPRPVTCHHVE---AAYADVSVDMGKANRLGAG--
MMAR_1987|M.marinum_M SGLETRDEFVVGSLAGPRPVTVHHAD---QTHADVTVDMGKANKLGPG--
MUL_3369|M.ulcerans_Agy99 SGLETRDEFVVGSLAGPRPVTVHHAD---QTHADVTVDMGKANKLGPG--
MAV_3619|M.avium_104 SGLETRDEFVVGSLAGPRPVTVHAAD---ATGADVSVDMGKANTLGSGGK
MLBr_00996|M.leprae_Br4923 SGLESCDEFVVGSLAGPRLVNVHHVD---ELNADVTVDMGKANLLGSGGP
Mvan_2435|M.vanbaalenii_PYR-1 SGLESRDEFVVGSLAGPRPVVLNALA---GVGADVTVEMGKANQLGVG--
Mflv_3961|M.gilvum_PYR-GCK GGLEHRSEFVVGSLAGPRPVVLHEAD---DTTADVTVEMGKAVRCGTG--
MAB_3043c|M.abscessus_ATCC_199 VGLEHRDEFVVASLAGPRPVRINSWS---QLTADVTVDMGPVKEFGAG--
*** .****.* **** * : *:*:*:** . * *
MSMEG_2735|M.smegmatis_MC2_155 --SAVVGGRSFDGLAVDVGNPHLACVG--ITADDLAALDVAAPVSFDPAL
TH_1519|M.thermoresistible__bu --TAVVGGRRFTGLAVDVGNPHLACVDPELTEDALAALDVAAPVQFDTEQ
Mb2745c|M.bovis_AF2122/97 --EAVVGGRRFHGLAVDVGNPHLACVDSQLTVDGLAALDVGAPVSFDGAQ
Rv2726c|M.tuberculosis_H37Rv --EAVVGGRRFHGLAVDVGNPHLACVDSQLTVDGLAALDVGAPVSFDGAQ
MMAR_1987|M.marinum_M --EAVVGGRRLSGVAVDVGNPHLACLDPGLTPEQLAALDVGAPVSFDHAQ
MUL_3369|M.ulcerans_Agy99 --EAVVGGRRLSGVAVDVGNPHLACLDPGLTPEQLAALDVGAPVSFDHAQ
MAV_3619|M.avium_104 AFEATVGGRRFAGLAVDVGNPHLACLDPELSVDELAALDVAAPVSFDAAQ
MLBr_00996|M.leprae_Br4923 AFAVTVGGRRFSGVAVDVGNPHLACMDPQLSLEELAALDLGAPVHLDRVQ
Mvan_2435|M.vanbaalenii_PYR-1 --AATVGGRQFNGLAVDVGNPHLACVDAAMSSRELAALDVAAPVVFDAAQ
Mflv_3961|M.gilvum_PYR-GCK --TATVGGRRFSGLAVDVGNPHLACVDEELTDAGLASLDVASPVAFDSTQ
MAB_3043c|M.abscessus_ATCC_199 --EAIVGGRRFSGLAIDVGNPHLACVDAVLTTEQLRILDVGAPVTFDEQL
. **** : *:*:*********:. :: * **:.:** :*
MSMEG_2735|M.smegmatis_MC2_155 FPDGVNVEVLTASVDG---AVSMRVHERGVGETRSCGTGTVAATVAALAH
TH_1519|M.thermoresistible__bu FPDGVNIEVLTPPVDS---VVSMRVHERGVGETRSCGTGTVAATVAALRH
Mb2745c|M.bovis_AF2122/97 FPDGVNVEVLTAPVDG---AVWMRVHERGVGETRSCGTGTVAAAVAALAA
Rv2726c|M.tuberculosis_H37Rv FPDGVNVEVLTAPVDG---AVWMRVHERGVGETRSCGTGTVAAAVAALAA
MMAR_1987|M.marinum_M FPEGVNVEVLTAPAAG---VVCMRVHERGVGETRSCGTGTVAAAVAALAS
MUL_3369|M.ulcerans_Agy99 FPEGVNVEVLTARAAG---VVCMRVHERGVGETRSCGTGTVAAAVAALAS
MAV_3619|M.avium_104 FPDGVNVEVLTAPAAG---VVHMRVHERGVGETRSCGTGTVAAAVAALTA
MLBr_00996|M.leprae_Br4923 FPDGVNIEVLTAPVDG---MVQMRVHERGVGETRSCGTGTVAAAVAALAS
Mvan_2435|M.vanbaalenii_PYR-1 FPDGVNVEVLTAPEDG---AVSMRVHERGVGETRSCGTGTVAAAVAALAH
Mflv_3961|M.gilvum_PYR-GCK FPHGVNVEILTAPRDG---AVAMRVHERGVGETRSCGTGTVAAAVAALDH
MAB_3043c|M.abscessus_ATCC_199 FPDGVNVEVLTAPSAGPANTVHMRVHERGVGETRSCGTGTVAAAYAALRH
**.***:*:**. . * *********************: ***
MSMEG_2735|M.smegmatis_MC2_155 DGADTGTLRVRIPGGEVTVTVTESTSYLRGPSVLVARGELDPHWWHA---
TH_1519|M.thermoresistible__bu LGADTGTMRVRVPGGTVTVTVTETTSHLRGPSVLVARGDISADWWAAQRD
Mb2745c|M.bovis_AF2122/97 VGSPTGTLTVHVPGGEVVVTVTDATSFLRGPSVLVARGDLADDWWNAMG-
Rv2726c|M.tuberculosis_H37Rv VGSPTGTLTVHVPGGEVVVTVTDATSFLRGPSVLVARGDLADDWWNAMG-
MMAR_1987|M.marinum_M AGQETGTLTVRIPGGDVTVNITDATSLLRGPSVLVASGEISEEWWRDQRR
MUL_3369|M.ulcerans_Agy99 AGQETGTLTVRIPGGDVTVNITDATSLLRGPSVLVASGEISEEWWRDQRR
MAV_3619|M.avium_104 AGADTGTLTVRVPGGDVVVTVTDATSYLRGPSVLVAHGEISEEWWQQAQR
MLBr_00996|M.leprae_Br4923 AGADTGTLTVRVPGGDVVITITDVTSYLRGPSVLVAHGELADAWWYSLAR
Mvan_2435|M.vanbaalenii_PYR-1 QGSQTGSLTVRIPGGEVVVTITDANSYLRGPSVLVAHGDLADEWWAAQG-
Mflv_3961|M.gilvum_PYR-GCK QGARTGTLRVRIPGGEVCVTVTETDSYLRGPSVLLAHGELAEQWWAAQH-
MAB_3043c|M.abscessus_ATCC_199 IGQRTGEVVVNIPGGQVRVTVTGESSFLRGPSMLLADGEISDEWWGGIGA
* ** : *.:*** * :.:* * *****:*:* *:: **
MSMEG_2735|M.smegmatis_MC2_155 -VAG-----------
TH_1519|M.thermoresistible__bu GMAGPAGALIEVHEQ
Mb2745c|M.bovis_AF2122/97 ---------------
Rv2726c|M.tuberculosis_H37Rv ---------------
MMAR_1987|M.marinum_M ---------------
MUL_3369|M.ulcerans_Agy99 ---------------
MAV_3619|M.avium_104 ---------------
MLBr_00996|M.leprae_Br4923 SC-------------
Mvan_2435|M.vanbaalenii_PYR-1 ---------------
Mflv_3961|M.gilvum_PYR-GCK ---------------
MAB_3043c|M.abscessus_ATCC_199 CG-------------