For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSSRPRLTSEPSDVPDGEQAQDPRTREEQAKNVCLRLLTVRARTRAELETQLTKRGYPDDVSARVLDRL TEVGLIDDEDFAEQWVRSRHLNAGKGKRALAVELRKKGVDDEVISSALADLDPAAERQRAEQLVRDKLRR ERLDGTPENDVKVTRRLVGMLARRGYNQSMAYDVVSVELANERERRRV
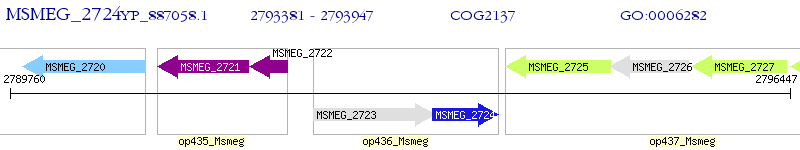
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2724 | recX | - | 100% (188) | recombination regulator RecX |
| M. smegmatis MC2 155 | MSMEG_5002 | - | 8e-06 | 28.11% (185) | hypothetical protein MSMEG_5002 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2755c | recX | 1e-53 | 64.81% (162) | recombination regulator RecX |
| M. gilvum PYR-GCK | Mflv_3974 | recX | 1e-66 | 70.68% (191) | recombination regulator RecX |
| M. tuberculosis H37Rv | Rv2736c | recX | 1e-53 | 64.81% (162) | recombination regulator RecX |
| M. leprae Br4923 | MLBr_00988 | recX | 1e-54 | 66.67% (162) | recombination regulator RecX |
| M. abscessus ATCC 19977 | MAB_3059c | recX | 2e-51 | 57.46% (181) | recombination regulator RecX |
| M. marinum M | MMAR_1978 | recX | 6e-56 | 63.93% (183) | regulatory protein RecX |
| M. avium 104 | MAV_3626 | recX | 2e-57 | 64.52% (186) | recombination regulator RecX |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3379 | recX | 4e-56 | 63.93% (183) | recombination regulator RecX |
| M. vanbaalenii PYR-1 | Mvan_2421 | recX | 2e-66 | 72.99% (174) | recombination regulator RecX |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3974|M.gilvum_PYR-GCK MTSPMTLFPLPSTSDSDGTPGTD--------------------EFATRAE
Mvan_2421|M.vanbaalenii_PYR-1 ----MTLFPLPSTSDP---AEAD--------------------ESTKRGE
MSMEG_2724|M.smegmatis_MC2_155 MTSSRPRLTSEPSDVPDG-EQAQ--------------------DPRTREE
Mb2755c|M.bovis_AF2122/97 MTVS---CPPPSTSE--------------------------------REE
Rv2736c|M.tuberculosis_H37Rv MTVS---CPPPSTSE--------------------------------REE
MMAR_1978|M.marinum_M MTVS---FPPPSTSEP------------------------------SREE
MUL_3379|M.ulcerans_Agy99 MTVS---FPPPSTSEP------------------------------SREE
MLBr_00988|M.leprae_Br4923 MTTS---CPPPSISD--------------------------------REE
MAV_3626|M.avium_104 MTAS---CPPPSTSEP------------------------------SREE
MAB_3059c|M.abscessus_ATCC_199 MTKS---SRPQSISDSVSVAGSQGTLDDLRARVASVPEAPTREPVDSRDE
. . * *
Mflv_3974|M.gilvum_PYR-GCK QARALCLRLLTVRARTRAELEAQLTKRGYPDDVSAEVLDRLAQVGLVDDE
Mvan_2421|M.vanbaalenii_PYR-1 QARALCLRLLTARARTRAELEAALAKRGYPDDIAAAVLDRLTQVGLVDDE
MSMEG_2724|M.smegmatis_MC2_155 QAKNVCLRLLTVRARTRAELETQLTKRGYPDDVSARVLDRLTEVGLIDDE
Mb2755c|M.bovis_AF2122/97 QARALCLRLLTARSRTRAELAGQLAKRGYPEDIGNRVLDRLAAVGLVDDT
Rv2736c|M.tuberculosis_H37Rv QARALCLRLLTARSRTRAELAGQLAKRGYPEDIGNRVLDRLAAVGLVDDT
MMAR_1978|M.marinum_M QARALCLRLLTARSRTRAELAGQLAKRGYPEDVGDRVLDRLTAVGLVDDA
MUL_3379|M.ulcerans_Agy99 QARALCLRLLTARSRTRAELAGQLAKRGYPEDVGDRVLDRLTAVGLVDDA
MLBr_00988|M.leprae_Br4923 QARILCLRLLTARSRTRAQLFGQLAKRGYADHVSERVLDRLAAVGLLDDN
MAV_3626|M.avium_104 QARALCLRLLTARARTRAELHGRLAKRGYPDDVSTAVLDRLAAVGLIDDA
MAB_3059c|M.abscessus_ATCC_199 QAWSYCLRLLTSRARTRAELTERLARRGYPDDVSERIMDRLATAGLVNDA
** ****** *:****:* *::***.:.:. ::***: .**::*
Mflv_3974|M.gilvum_PYR-GCK DFAEQWVRSRRINAGKGKRALASELRNKGVDSEVIDAALADIDAGAERTR
Mvan_2421|M.vanbaalenii_PYR-1 DFAEQWVRSRRLNAGKGKRALAAELRKKGVDNEVIDGALAGIDAGAERTR
MSMEG_2724|M.smegmatis_MC2_155 DFAEQWVRSRHLNAGKGKRALAVELRKKGVDDEVISSALADLDPAAERQR
Mb2755c|M.bovis_AF2122/97 DFAEQWVQSRRANAAKSKRALAAELHAKGVDDDVITTVLGGIDAGAERGR
Rv2736c|M.tuberculosis_H37Rv DFAEQWVQSRRANAAKSKRALAAELHAKGVDDDVITTVLGGIDAGAERGR
MMAR_1978|M.marinum_M DFAADWVQSRRANAGKSRRALAAELQAKGVDQDVIGTALAGLDAGAERGR
MUL_3379|M.ulcerans_Agy99 DFAADWVQSRRANAGKSRRALAAELQAKGVDQDVIGTALAGLDAGAERGR
MLBr_00988|M.leprae_Br4923 DFAEQWVRSRRANVGKSKRALAADLRAKGIDSEVITTVLAGIDPAVERER
MAV_3626|M.avium_104 DFAQQWVQSRRAHAGKSKRALAAELHTKGVDNDVITSVLAGIDAGAERDR
MAB_3059c|M.abscessus_ATCC_199 DFATQWVQSRHTYSGKGKRALAAELRTKGVSAENAAAALAQIDGEAERSR
*** :**:**: .*.:**** :*: **:. : .*. :* .** *
Mflv_3974|M.gilvum_PYR-GCK AEQLVRDKLRREKLGDPGDRDAENKVMRRLVGMLARRGYHQSMALDVVTT
Mvan_2421|M.vanbaalenii_PYR-1 AEQLVRDKLRREKLGDPDDRDAENKVARRLVGMLARRGYNQTMAFDVVTA
MSMEG_2724|M.smegmatis_MC2_155 AEQLVRDKLRRERLD--GTPENDVKVTRRLVGMLARRGYNQSMAYDVVSV
Mb2755c|M.bovis_AF2122/97 AEKLVRARLRREVLIDDGTD--EARVSRRLVAMLARRGYGQTLACEVVIA
Rv2736c|M.tuberculosis_H37Rv AEKLVRARLRREVLIDDGTD--EARVSRRLVAMLARRGYGQTLACEVVIA
MMAR_1978|M.marinum_M AEQLVRTKLRREKLTED-----DARVTRRLVAMLARRGYSQTVACEVVIA
MUL_3379|M.ulcerans_Agy99 AEQLVRTKLRREKLTED-----DARVTRRLVAMLARRGYSQTVACEVVIA
MLBr_00988|M.leprae_Br4923 AEQLVRTRLRREVLSED-----DARVSRRLMAMLARRGYSQTTICEVVVA
MAV_3626|M.avium_104 AEQLVRSKLRRETLAQD-----DARVTRRLVGMLARRGYSQNLACEVVLA
MAB_3059c|M.abscessus_ATCC_199 AAELVTKKLRSENLDDG-----GIKAARRLVAMLARRGYGQSMAYDVVKN
* :** :** * * :. ***:.******* *. :**
Mflv_3974|M.gilvum_PYR-GCK ELANERERRTV-
Mvan_2421|M.vanbaalenii_PYR-1 ELANERERRKV-
MSMEG_2724|M.smegmatis_MC2_155 ELANERERRRV-
Mb2755c|M.bovis_AF2122/97 ELAAERERRRV-
Rv2736c|M.tuberculosis_H37Rv ELAAERERRRV-
MMAR_1978|M.marinum_M ELAAERDRRRV-
MUL_3379|M.ulcerans_Agy99 ELAAERDRRRV-
MLBr_00988|M.leprae_Br4923 ELAAERERRRV-
MAV_3626|M.avium_104 ELAAERERRRV-
MAB_3059c|M.abscessus_ATCC_199 ALASEKDRRDVG
** *::** *