For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAEIAPLRVQLIAKTEFMAPPGVPWETDADGGEALTEFAGRACYQSWSKPNPKTATNAGYISHIIDVGHF SVLEHASVSFYISGISRSATHELIRHRHFSYSQLSQRYVPEHDSEVVVPPGFEDDPELVEIFTSAVDASR ATYTELLKRLEAKFADQPNAVLRRKQARQAARAVLPNATETRIVVTGNYRAWRHFIAMRASEHADVEIRR LAIACLRELVTVAPAVFSDFVISTLADGSEVATSPLATEA
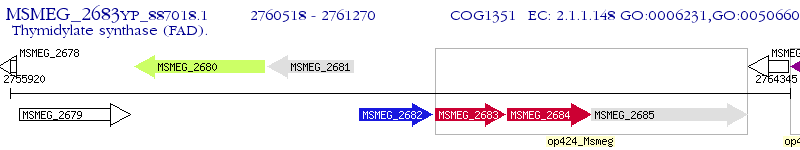
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2683 | thyX | - | 100% (250) | FAD-dependent thymidylate synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2775c | thyX | 1e-123 | 86.00% (250) | FAD-dependent thymidylate synthase |
| M. gilvum PYR-GCK | Mflv_4001 | thyX | 1e-120 | 85.14% (249) | FAD-dependent thymidylate synthase |
| M. tuberculosis H37Rv | Rv2754c | thyX | 1e-123 | 86.00% (250) | FAD-dependent thymidylate synthase |
| M. leprae Br4923 | MLBr_01514 | thyX | 1e-114 | 81.10% (254) | FAD-dependent thymidylate synthase |
| M. abscessus ATCC 19977 | MAB_3085c | thyX | 1e-112 | 77.91% (249) | FAD-dependent thymidylate synthase |
| M. marinum M | MMAR_1961 | thyX | 1e-126 | 88.76% (249) | thymidylate synthase ThyX |
| M. avium 104 | MAV_3645 | thyX | 1e-126 | 88.40% (250) | FAD-dependent thymidylate synthase |
| M. thermoresistible (build 8) | TH_0835 | - | 1e-121 | 85.20% (250) | PROBABLE THYMIDYLATE SYNTHASE THYX (TS) (TSase) |
| M. ulcerans Agy99 | MUL_2183 | thyX | 1e-125 | 88.35% (249) | FAD-dependent thymidylate synthase |
| M. vanbaalenii PYR-1 | Mvan_2386 | thyX | 1e-124 | 86.75% (249) | FAD-dependent thymidylate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4001|M.gilvum_PYR-GCK ---MAEIAPLRVQLIAKTEFSAPPDVEWSTDADGGAALVEFAGRACYQSW
Mvan_2386|M.vanbaalenii_PYR-1 ---MAEIAPLRVQLIAKTEFSAPPDVPWSTDADGGAALVEFAGRACYQSW
Mb2775c|M.bovis_AF2122/97 ---MAETAPLRVQLIAKTDFLAPPDVPWTTDADGGPALVEFAGRACYQSW
Rv2754c|M.tuberculosis_H37Rv ---MAETAPLRVQLIAKTDFLAPPDVPWTTDADGGPALVEFAGRACYQSW
MMAR_1961|M.marinum_M ---MAETAPLRVQLIAKTEFLAPPDVPWTTDADGGEALVEFAGRACYQSW
MUL_2183|M.ulcerans_Agy99 MSAVAETAPLRVQLIAKTEFLAPPDVPWTTDADGGEALVEFAGRACYQSW
MAV_3645|M.avium_104 ---MAEIAPLRVQLIAKTDFLAPPDVPWSTDADGGPALVEFAGRACYQSW
MLBr_01514|M.leprae_Br4923 ---MAQIAPLRVQLIAKTEFLAPPDVSWTTDADGGSALVEFAGRACYQSW
MSMEG_2683|M.smegmatis_MC2_155 ---MAEIAPLRVQLIAKTEFMAPPGVPWETDADGGEALTEFAGRACYQSW
TH_0835|M.thermoresistible__bu ---VAEIAPLRVQLIAKTEFLAPPDVPWSTDADGGEALVEFAGRACYQSW
MAB_3085c|M.abscessus_ATCC_199 ---MAEIVPLRVQLIAKTDFIAPPDIDWSTDADGGPALVEFAGRACYQSW
:*: .**********:* ***.: * ****** **.***********
Mflv_4001|M.gilvum_PYR-GCK SKPNPRTATNATYVRHIIDVGHFSVLEHASVSFYITGLSRSCTHELIRHR
Mvan_2386|M.vanbaalenii_PYR-1 DKPNPRTATNAGYVRHIIDVGHFSVLEHASVSFYITGISRSCTHELIRHR
Mb2775c|M.bovis_AF2122/97 SKPNPKTATNAGYLRHIIDVGHFSVLEHASVSFYITGISRSCTHELIRHR
Rv2754c|M.tuberculosis_H37Rv SKPNPKTATNAGYLRHIIDVGHFSVLEHASVSFYITGISRSCTHELIRHR
MMAR_1961|M.marinum_M SKPNPKTATNAGYLRHIIDVGHFAVLEHASVSFYISGISRSCTHELIRHR
MUL_2183|M.ulcerans_Agy99 SKPNPKTATNAGYLRHIIDVGHFAVLEHASVSFYISGISRSCTHELIRHR
MAV_3645|M.avium_104 SKPNPKTATNAGYIKHIIDVGHFSVLEHASVSFYITGISRSCTHELIRHR
MLBr_01514|M.leprae_Br4923 SKPNPRTATNAAYIKHIIDVGHVAVLEHASVSFYISGISRSCTHELIRHR
MSMEG_2683|M.smegmatis_MC2_155 SKPNPKTATNAGYISHIIDVGHFSVLEHASVSFYISGISRSATHELIRHR
TH_0835|M.thermoresistible__bu SKPNPKTATNAAYIRHIIDVGHLSVLEHASVSFYITGISRSCTHELIRHR
MAB_3085c|M.abscessus_ATCC_199 SKPNPRTATNESYLRHVIEVGHLSVLEHASATFYITGISRSCTHELIRHR
.****:**** *: *:*:***.:******.:***:*:***.********
Mflv_4001|M.gilvum_PYR-GCK HFSYSQLSQRYVPENDAEVVAPPGIEDDPELLALFTAATDASRAAYTELL
Mvan_2386|M.vanbaalenii_PYR-1 HFSYSQLSQRYVPEHDSQVVAPPGIEDDPELLEIFTEAADAGRAAYTELL
Mb2775c|M.bovis_AF2122/97 HFSYSQLSQRYVPEKDSRVVVPPGMEDDADLRHILTEAADAARATYSELL
Rv2754c|M.tuberculosis_H37Rv HFSYSQLSQRYVPEKDSRVVVPPGMEDDADLRHILTEAADAARATYSELL
MMAR_1961|M.marinum_M HFSYSQLSQRYVPEGDSRVVVPPGLEDDPELREILIAAADASRATYTELL
MUL_2183|M.ulcerans_Agy99 HFSYSQLSQRYVPEGDSRVVVPPGLEDDPELREILIAAADASRATYIELL
MAV_3645|M.avium_104 HFSYSQLSQRYVPENDSRVVVPPGLDDDPELQQILAAAADASRATYTELL
MLBr_01514|M.leprae_Br4923 HFSYSQLSQRYVPEKDAQVVVPPDMEDDDELQQILIAAVEASRATYTELL
MSMEG_2683|M.smegmatis_MC2_155 HFSYSQLSQRYVPEHDSEVVVPPGFEDDPELVEIFTSAVDASRATYTELL
TH_0835|M.thermoresistible__bu HFSYSQLSQRYVPDGDARVVLPPGLEDDPELQEIFTAAADAGHAAYTELL
MAB_3085c|M.abscessus_ATCC_199 HFSYSQLSQRFVPEDDSNVVLPPAVEDDPELVALVRKATDASRAAYVELL
**********:**: *:.** ** .:** :* :. *.:*.:*:* ***
Mflv_4001|M.gilvum_PYR-GCK NRLEAKLAD----QGTSTLRRKQARQAARAVLPNATETRIVVTGNYRAWR
Mvan_2386|M.vanbaalenii_PYR-1 TRLEARLAD----QPNATLRRKQARQAARAVLPNATETRIVVTGNYRAWR
Mb2775c|M.bovis_AF2122/97 AKLEAKFAD----QPNAILRRKQARQAARAVLPNATETRIVVTGNYRAWR
Rv2754c|M.tuberculosis_H37Rv AKLEAKFAD----QPNAILRRKQARQAARAVLPNATETRIVVTGNYRAWR
MMAR_1961|M.marinum_M TKLEARFAD----QPNAVLRRKQARQAARAVLPNATETRIVVSGNYRAWR
MUL_2183|M.ulcerans_Agy99 TKLEARFAD----QPNAVLRRKQARQAARAVLPNATETRIVVSGNYRAWR
MAV_3645|M.avium_104 ARLEAKFAD----QPSAVLRRKQARQAARAVLPNATETRIVVTGNYRAWR
MLBr_01514|M.leprae_Br4923 VKLNAKLMAGELGGNRAVLRRKQARQAAHAVLPNANETRIVVTGNYRAWR
MSMEG_2683|M.smegmatis_MC2_155 KRLEAKFAD----QPNAVLRRKQARQAARAVLPNATETRIVVTGNYRAWR
TH_0835|M.thermoresistible__bu TRLQAALAD----QPNAVLRRKQARQAARAVLPNATETRIVVTGNYRAWR
MAB_3085c|M.abscessus_ATCC_199 EKLESTLAD----VPNAVLRRKQARQAARSVLPNATETRIVVTGNYRAWR
:*:: : : **********::*****.******:*******
Mflv_4001|M.gilvum_PYR-GCK HFIAMRASEHADVEIRRLAIECLRRLVAVAPQVFSDFEITALADGTEVAT
Mvan_2386|M.vanbaalenii_PYR-1 HFIAMRASEHADLEIRRLAIECLRQLVDAAPQVFSDFEIVVLADGTEVAT
Mb2775c|M.bovis_AF2122/97 HFIAMRASEHADVEIRRLAIECLRQLAAVAPAVFADFEVTTLADGTEVAT
Rv2754c|M.tuberculosis_H37Rv HFIAMRASEHADVEIRRLAIECLRQLAAVAPAVFADFEVTTLADGTEVAT
MMAR_1961|M.marinum_M HFIAMRASEHADVEIRRLAIECLRQLVAVAPAVFADFEVTTLADGSEVAT
MUL_2183|M.ulcerans_Agy99 HFIAMRASEHADVEIRRLAIECLRQLVAVAPAVFADFEVTTLADGSEVAT
MAV_3645|M.avium_104 HFIAMRASEHADVEIRRLAIECLRRLADVAPAVFADFEIATLADGTEVAT
MLBr_01514|M.leprae_Br4923 HFIAMRASEHADVEIRRLAIVCLRRLVDVAPAVFADFEITALADGTEVAT
MSMEG_2683|M.smegmatis_MC2_155 HFIAMRASEHADVEIRRLAIACLRELVTVAPAVFSDFVISTLADGSEVAT
TH_0835|M.thermoresistible__bu HFIAVRASEQADLEIRRLAIECLRRLTEVAPAAFADFEIATLADGTEVAT
MAB_3085c|M.abscessus_ATCC_199 HFIAMRASEHADVEIRRLAIACLRQLADLAPSIFGDFDIATLADGTEVAI
****:****:**:******* ***.*. ** *.** : .****:***
Mflv_4001|M.gilvum_PYR-GCK SPLATEV
Mvan_2386|M.vanbaalenii_PYR-1 SPLATEV
Mb2775c|M.bovis_AF2122/97 SPLATEA
Rv2754c|M.tuberculosis_H37Rv SPLATEA
MMAR_1961|M.marinum_M SPLATEV
MUL_2183|M.ulcerans_Agy99 SPLATEV
MAV_3645|M.avium_104 SPLATEA
MLBr_01514|M.leprae_Br4923 SPLATEA
MSMEG_2683|M.smegmatis_MC2_155 SPLATEA
TH_0835|M.thermoresistible__bu SPLATEA
MAB_3085c|M.abscessus_ATCC_199 SPLVYEG
***. *