For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPGIAELALGAAPIAGGAMLGVIAGNLRPPDVRGMIAKDMDLLERLPEEEVELRDRLRASINQRISDLVT AGERSREIRLAATSYQGNWRDIVVFLCAVLFTIVWWNIDHDRTNWLPMFVIMIVVSVVAGIYAGRGILRA LQTFARRNSDKPQ
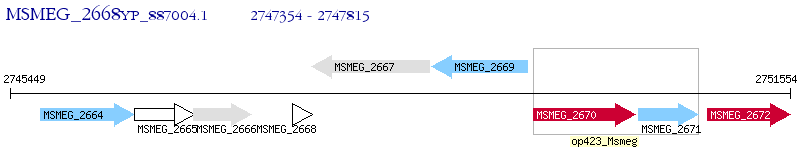
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2668 | - | - | 100% (153) | hypothetical protein MSMEG_2668 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4010 | - | 8e-54 | 64.58% (144) | hypothetical protein Mflv_4010 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3093c | - | 4e-49 | 57.82% (147) | hypothetical protein MAB_3093c |
| M. marinum M | MMAR_2637 | - | 3e-46 | 54.30% (151) | hypothetical protein MMAR_2637 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0694 | - | 4e-56 | 67.33% (150) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2377 | - | 3e-52 | 58.17% (153) | hypothetical protein Mvan_2377 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2668|M.smegmatis_MC2_155 MPGIAELALGAAPIAGGAMLGVIAGNLRPPDVRGMIAKDMDLLERLPEEE
TH_0694|M.thermoresistible__bu MPGIAELALGAAPVAGGALLALAAGNLKGPDFRGMIAKDMDLLERIPVED
Mflv_4010|M.gilvum_PYR-GCK MPGMAEFALAGAPIAGGALLGLAAGNLRPPDVRDAIAKDLDLLERLPAEQ
Mvan_2377|M.vanbaalenii_PYR-1 MPGMAEFALAGAPIAGGALLGLAAGNLRPPDVRDAIARDLDLMERLPADQ
MAB_3093c|M.abscessus_ATCC_199 MPALAELALGAAPIAGGALLGAVAGNLKPPDIRELISKDFDLLDRIPEEQ
MMAR_2637|M.marinum_M MVGLAEIALNTAPIFGGALLAVSAGQFKRPDYRAIIKQDMDLLDRLPPDA
* .:**:** **: ***:*. **::: ** * * :*:**::*:* :
MSMEG_2668|M.smegmatis_MC2_155 VELRDRLRASINQRISDLVTAGERSREIRLAATSYQGNWRDIVVFLCAVL
TH_0694|M.thermoresistible__bu VERRARLQASINQRIDDLIATTERSRALREAAMSYGGNWRDIVLFICAVL
Mflv_4010|M.gilvum_PYR-GCK VERRAELQRIIDMRIDGLVAAADKSRELRELAMSYEGNWRDIVLFVCAIM
Mvan_2377|M.vanbaalenii_PYR-1 AERRAELQRIIDMRIDGLIAATDKSRELRELAMSYQGNWRDIVVFICAIL
MAB_3093c|M.abscessus_ATCC_199 AERRAAVRRMVDQRIDDLVASNDRGRSLREAALSYGGGWRDILVFVGAVL
MMAR_2637|M.marinum_M TERRADLQRTIDSRIDDLVNISQRNRALRQAAASYQGNWRDLVLLLCVVL
.* * :: :: **..*: ::.* :* * ** *.***::::: .::
MSMEG_2668|M.smegmatis_MC2_155 FTIVWWNIDHDRTNWLPMFVIMIVVSVVAGIYAGRGILR---ALQTFARR
TH_0694|M.thermoresistible__bu FTLVWWHVDHDRSNWLVMFVVLIGFCVLTALYAVRGILR---AFGSLRRN
Mflv_4010|M.gilvum_PYR-GCK FTLVWWHVDHSRSNWLVMFVVMIVLTVIVGIYAARGVLR---GLASFLAN
Mvan_2377|M.vanbaalenii_PYR-1 FTIVWWHVEHTRSNWLLMFILLLVLTVIVGIYAARGVMR---AVASFLAR
MAB_3093c|M.abscessus_ATCC_199 FTVIWWNIEHTRSNWLVMFLVMLALTLIAGFYAVRTVSRGLYGAAGRLRH
MMAR_2637|M.marinum_M FTFVWWGVSHSRANWLPTFIALIVLSCVAAVYAGRGVLR---AITSLLAR
**.:** :.* *:*** *: :: . :...** * : * . .
MSMEG_2668|M.smegmatis_MC2_155 NSDKPQ
TH_0694|M.thermoresistible__bu RPD---
Mflv_4010|M.gilvum_PYR-GCK RRRGRQ
Mvan_2377|M.vanbaalenii_PYR-1 RRRQAQ
MAB_3093c|M.abscessus_ATCC_199 RSPGSS
MMAR_2637|M.marinum_M GQDQA-