For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDSLLSLVLPDRVASAEVYDDPPGLSPLPEEEPLIARSVAKRRNEFVTVRYCARQALGELGVGPVPILK GDKGEPCWPDGVVGSLTHCQGFRGAVVGRSTDVRSVGIDAEPHDVLPNGVLDAITLPIERAELRGLPGDL HWDRILFCAKEATYKAWYPLTHRWLGFEDAHITFEVDGSGTAGSFRSRILIDPVAEHGPPLTALDGRWSV RDGLAVTAIVL
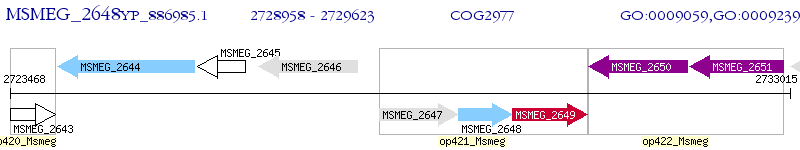
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2648 | - | - | 100% (221) | Sfp-type phosphopantetheinyl transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2817c | - | 2e-93 | 74.89% (223) | hypothetical protein Mb2817c |
| M. gilvum PYR-GCK | Mflv_4029 | - | 1e-104 | 80.00% (220) | 4'-phosphopantetheinyl transferase |
| M. tuberculosis H37Rv | Rv2794c | - | 6e-93 | 74.44% (223) | hypothetical protein Rv2794c |
| M. leprae Br4923 | MLBr_01547 | - | 7e-93 | 74.32% (222) | hypothetical protein MLBr_01547 |
| M. abscessus ATCC 19977 | MAB_3117c | - | 9e-89 | 68.78% (221) | 4'-phosphopantetheinyl transferase |
| M. marinum M | MMAR_1916 | pptII | 1e-96 | 77.48% (222) | phosphopantetheinyl transferase, PptII |
| M. avium 104 | MAV_3683 | - | 1e-96 | 77.33% (225) | Sfp-type phosphopantetheinyl transferase |
| M. thermoresistible (build 8) | TH_1427 | - | 1e-96 | 76.47% (221) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2141 | pptII | 2e-95 | 76.58% (222) | phosphopantetheinyl transferase, PptII |
| M. vanbaalenii PYR-1 | Mvan_2328 | - | 1e-97 | 74.55% (224) | 4'-phosphopantetheinyl transferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4029|M.gilvum_PYR-GCK -MIAAPLLSGVLPG-EVDALAAAEMYTDPRELAPLPEEEPLIAKSVAKRR
Mvan_2328|M.vanbaalenii_PYR-1 -MIAATLLAGVLPG-EIDALAAAEMYSDPKELAPLPEEEPLIARSVAKRR
MSMEG_2648|M.smegmatis_MC2_155 --MTDSLLSLVLP----DRVASAEVYDDPPGLSPLPEEEPLIARSVAKRR
TH_1427|M.thermoresistible__bu -VRVTTLLGRLVP----ATVAAAELYDDPPGVAPLPEEEPLIARSVDKRR
MMAR_1916|M.marinum_M -MTISMLVSSVLPGTVVDDLAYAELYSDPPGLVPLPEEEPLIARSVAKRR
MUL_2141|M.ulcerans_Agy99 -MTISMLVSSVLPGTVVDDLAYAELYSDPPGLVPLPEEEPLIAKSVAKRR
Mb2817c|M.bovis_AF2122/97 -MTVGTLVASVLPATVFEDLAYAELYSDPPGLTPLPEEAPLIARSVAKRR
Rv2794c|M.tuberculosis_H37Rv -MTVGTLVASVLPATVFEDLAYAELYSDPPGLTPLPEEAPLIARSVAKRR
MLBr_01547|M.leprae_Br4923 -MTVSMLVSSVLPDYASQDLEYAELYSDPPGLTPLPEEELLIAKSVAKRR
MAV_3683|M.avium_104 --MTGTLVSSVLP--ASDGLAYSEVYSDPPGLAPLPEEEPLIARSVAKRR
MAB_3117c|M.abscessus_ATCC_199 MPVTDQLIASVVP----ELLPSAELYEDPPGLEPLPEEEPLIAKSVAKRR
*:. ::* : :*:* ** : ***** ***:** ***
Mflv_4029|M.gilvum_PYR-GCK NEFITVRYCARQALVDLGMEPVPILKGDKGEPCWPDGVVGSLTHCEGFRG
Mvan_2328|M.vanbaalenii_PYR-1 NEFITVRFCARQALVDLGMEPVPILKGDKGEPCWPDGIVGSLTHCEGFRG
MSMEG_2648|M.smegmatis_MC2_155 NEFVTVRYCARQALGELGVGPVPILKGDKGEPCWPDGVVGSLTHCQGFRG
TH_1427|M.thermoresistible__bu NEFVTVRHCARLALGELGVPAVPILKGEKGEPRWPDGVVGSLTHCAGYRG
MMAR_1916|M.marinum_M NEFITVRHCARVALGDLGVPPVPILKGDKGQPCWPDGVVGSLTHCSGYRG
MUL_2141|M.ulcerans_Agy99 NEFITVRHCARVALGDLGVPPVPILKGDKGQPCWPDGVVGSLTHCSGYRG
Mb2817c|M.bovis_AF2122/97 NEFITVRHCARIALDQLGVPPAPILKGDKGEPCWPDGVVGSLTHCAGYRG
Rv2794c|M.tuberculosis_H37Rv NEFITVRHCARIALDQLGVPPAPILKGDKGEPCWPDGMVGSLTHCAGYRG
MLBr_01547|M.leprae_Br4923 NEFITARYCARIALGRLRVPPVPILKGDKGEPCWPDGVVGSLTHCSGYRG
MAV_3683|M.avium_104 NEFITVRHCARIALGELGLPPAPILKGEKGEPRWPDGVVGSLTHCTGYRG
MAB_3117c|M.abscessus_ATCC_199 NEFITVRYCARQALSVLGIPEVPILKGDKGQPLWPDGIVGSMTHTEGFRG
***:*.*.*** ** * : .*****:**:* ****:***:** *:**
Mflv_4029|M.gilvum_PYR-GCK AAIGRRSDVRSLGIDAEPHDVLPAGVLDAISLPVERHELG-GMPGGVHWD
Mvan_2328|M.vanbaalenii_PYR-1 AAVARRAEVRSVGIDAEPHDVLPTGVLDAISLPVERHELG-DMPTGVHWD
MSMEG_2648|M.smegmatis_MC2_155 AVVGRSTDVRSVGIDAEPHDVLPNGVLDAITLPIERAELR-GLPGDLHWD
TH_1427|M.thermoresistible__bu AAVARSGEVRSVGIDAEPHDVLPRGVLDAVSLPAERREIS-ALPDGLHWD
MMAR_1916|M.marinum_M AVVGRSAAVRSVGIDAEPHDVLPNGVLDAISLPEERDEIPSAMPDGLHWD
MUL_2141|M.ulcerans_Agy99 AVVGRSAAVRSVGIDAEPHDVLSNGVLDAISLPEERDEIPSAMPDGLHWD
Mb2817c|M.bovis_AF2122/97 AVVGRRDAVRSVGIDAEPHDVLPNGVLDAISLPAERADMPRTMPAALHWD
Rv2794c|M.tuberculosis_H37Rv AVVGRRDAVRSVGIDAEPHDVLPNGVLDAISLPAERADMPRTMPAALHWD
MLBr_01547|M.leprae_Br4923 AVVGRSAAVRSVGIDAEPHEMLPNGVLDVISLPEERSEMRRKLPSVLYWD
MAV_3683|M.avium_104 AVVGRTGAVRSVGIDAEPHDVLPDGVLNAISLPAERSEIPSALPGDLHWD
MAB_3117c|M.abscessus_ATCC_199 AVVGRTGEVRSVGIDAEPHDVLPNGVLKSIALPVERDELD-ALPAGTHWD
*.:.* ***:*******::*. ***. ::** ** :: :* :**
Mflv_4029|M.gilvum_PYR-GCK RVLFCAKEATYKAWYPLTHRWLGFEDAHITFDVDS---TGQAGTFRSRIL
Mvan_2328|M.vanbaalenii_PYR-1 RVLFCAKEATYKAWFPLTHRWLGFEDAHITFDIDSSDNSGQSGTFTSQIL
MSMEG_2648|M.smegmatis_MC2_155 RILFCAKEATYKAWYPLTHRWLGFEDAHITFEVDGS---GTAGSFRSRIL
TH_1427|M.thermoresistible__bu RILFCAKEATYKAWFPLTRRWLGFEDAHITFEVEEVSAEGASGTFRSRIL
MMAR_1916|M.marinum_M RILFCAKEATYKVWFPLTNRWLGFEDAHITFEADDS--GRT-GRFVSRIL
MUL_2141|M.ulcerans_Agy99 RILFCAKEATYKVWFPLTNRWLGFEDAHITFEADDS--GRT-GRFVSRIL
Mb2817c|M.bovis_AF2122/97 RILFCAKEATYKAWFPLTKRWLGFEDAHITFETDST--GWT-GRFVSRIL
Rv2794c|M.tuberculosis_H37Rv RILFCAKEATYKAWFPLTKRWLGFEDAHITFETDST--GWT-GRFVSRIL
MLBr_01547|M.leprae_Br4923 RILFCAKEATYKAWFPLTKRWLGFEDAHITFDVDNL--GSS-GGFVSRIL
MAV_3683|M.avium_104 RILFCAKEATYKAWFPLTRRWLGFEDAHITFEADHP--GATTGGFVSRIL
MAB_3117c|M.abscessus_ATCC_199 RLLFCAKETTYKAWFPLTARWLGFEDAHITIDPDGT--------FTSRIL
*:******:***.*:*** ***********:: : * *:**
Mflv_4029|M.gilvum_PYR-GCK IDPAAESGPPLTALAGRWSVRNGIALTAIVL-
Mvan_2328|M.vanbaalenii_PYR-1 IDPEAESGPPLTSLAGRWSVRNGIALTAIVL-
MSMEG_2648|M.smegmatis_MC2_155 IDPVAEHGPPLTALDGRWSVRDGLAVTAIVL-
TH_1427|M.thermoresistible__bu IDPRALSGPPLRTLPGRWSVRNGLAVTAIVL-
MMAR_1916|M.marinum_M IDPSALWGPPLTTLHGRWSVERGLVLTAIVL-
MUL_2141|M.ulcerans_Agy99 IDPSALWGPPLTTLHGRWSVERGLVLTAIVL-
Mb2817c|M.bovis_AF2122/97 IDGSTLSGPPLTTLRGRWSVERGLVLTAIVL-
Rv2794c|M.tuberculosis_H37Rv IDGSTLSGPPLTTLRGRWSVERGLVLTAIVL-
MLBr_01547|M.leprae_Br4923 VDGSALSGPPLTVLTGRWSVDRGLVLTAIVL-
MAV_3683|M.avium_104 IDPAALCGPPLTALSGRWSVARGLVLTAIVL-
MAB_3117c|M.abscessus_ATCC_199 VDGRANDGTVLSAFDGRWIIDKGLILTAIVVP
:* : *. * : *** : *: :****: