For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VIQRETPASTRRRIPDTPVGPVRTCIGCRKRELAADLLRTVAVRDGNGNYSVSVDTSASMPGRGAWLHPD PKCAELAVRRRAFARALRIAGSPDTSGVNEYFESLGTPEHRGIENR
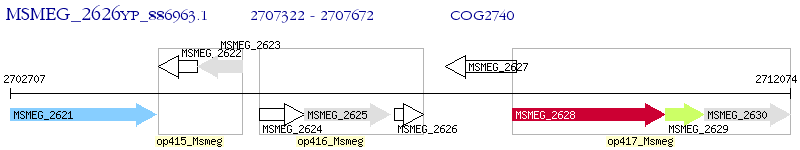
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2626 | - | - | 100% (116) | hypothetical protein MSMEG_2626 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2865c | - | 1e-33 | 72.53% (91) | hypothetical protein Mb2865c |
| M. gilvum PYR-GCK | Mflv_4040 | - | 5e-32 | 55.83% (120) | hypothetical protein Mflv_4040 |
| M. tuberculosis H37Rv | Rv2840c | - | 1e-33 | 72.53% (91) | hypothetical protein Rv2840c |
| M. leprae Br4923 | MLBr_01557 | - | 2e-29 | 62.64% (91) | hypothetical protein MLBr_01557 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1893 | - | 9e-35 | 62.96% (108) | hypothetical protein MMAR_1893 |
| M. avium 104 | MAV_3694 | - | 2e-22 | 60.81% (74) | hypothetical protein MAV_3694 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2121 | - | 6e-23 | 61.84% (76) | hypothetical protein MUL_2121 |
| M. vanbaalenii PYR-1 | Mvan_2313 | - | 3e-32 | 57.72% (123) | hypothetical protein Mvan_2313 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2865c|M.bovis_AF2122/97 ---------------------MRTCVGCRKRGLAVELLRVVAVSTGNG--
Rv2840c|M.tuberculosis_H37Rv ---------------------MRTCVGCRKRGLAVELLRVVAVSTGNG--
MMAR_1893|M.marinum_M -MIQRKPSAMAHGRPDIPCGPVRTCVGCRKRELAVELLRVVAESTGNG--
MUL_2121|M.ulcerans_Agy99 --------------------------------MAVELLRVVAESTGNG--
MLBr_01557|M.leprae_Br4923 ---------------------MRTCVGCRKRELAVELLRVVAPSTGKG--
MAV_3694|M.avium_104 ---------------------------------------MVAVPTGNG--
Mflv_4040|M.gilvum_PYR-GCK MIQRETSARMRESTINPFAGPVRTCIGCRRRELAVELLRLVAVDSANGSG
Mvan_2313|M.vanbaalenii_PYR-1 MIQRETSARMRKSAINPTAGPVRTCIGCRKRELAVELLRVVAADSGNGPG
MSMEG_2626|M.smegmatis_MC2_155 MIQRETPASTRRRIPDTPVGPVRTCIGCRKRELAADLLRTVAVRDGNG--
** .:*
Mb2865c|M.bovis_AF2122/97 --NYAVIVDTATSLPGRGAWLHPLRQCAQQAIRRRAFARALRIAGSPDTS
Rv2840c|M.tuberculosis_H37Rv --NYAVIVDTATSLPGRGAWLHPLRQCAQQAIRRRAFARALRIAGSPDTS
MMAR_1893|M.marinum_M --NYAVIVDTARGLPGRGAWLHPVPQCVQQAIRRRAFTRALRITGSPDIS
MUL_2121|M.ulcerans_Agy99 --NYAVIVDTARGLPGRGAWLHPVPQCVQQAIRRRAFTRALRITGSPDIS
MLBr_01557|M.leprae_Br4923 --SYAVIVDTASSLSGRGAWLHPDMQCVQQAIRRRAFTGALRIAGSPDTS
MAV_3694|M.avium_104 --EFAAIVDTAGNLPGRGAWLHPAPQCAQQAIRRRAFTKALRITGSPDTS
Mflv_4040|M.gilvum_PYR-GCK NRSSAVTVDAARKLPGRGAWLHPDPGCLQAAIRRRAFGRALRITGSPDIT
Mvan_2313|M.vanbaalenii_PYR-1 NGPSTVTVDSARRLPGRGAWLHPDPVCLQAAVRRRAFGRALRITGSPDIT
MSMEG_2626|M.smegmatis_MC2_155 --NYSVSVDTSASMPGRGAWLHPDPKCAELAVRRRAFARALRIAGSPDTS
:. **:: :.******** * : *:***** ****:**** :
Mb2865c|M.bovis_AF2122/97 AVVEYLESLGELEPPGNRTGSNRT-------
Rv2840c|M.tuberculosis_H37Rv AVVEYLESLGELEPPGNRTGSNRT-------
MMAR_1893|M.marinum_M AVVEHIGGLDVPGPPSNRTGSKEHEHTVKSR
MUL_2121|M.ulcerans_Agy99 AVVEHIGGLDVPGPPSNRTGSKEHEHTVKSR
MLBr_01557|M.leprae_Br4923 AVVEHIEFLSELDRPGNRTGSKEHEHTVKSR
MAV_3694|M.avium_104 AVVEHIESLHAPERPATEQVAKNMSTP----
Mflv_4040|M.gilvum_PYR-GCK AVSEHLLPQEQPEAPGQENR-----------
Mvan_2313|M.vanbaalenii_PYR-1 ALSERFPTGDAHGTPGQENR-----------
MSMEG_2626|M.smegmatis_MC2_155 GVNEYFESLGTPEHRGIENR-----------
.: * : . .