For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PTLLWFRRDLRCADHPAVLDAAQGDADVLACYVLDPRLTGSSGDRRLAYLYGALRDLREQLGGKLLVTRG RPEQRIPELAAAIGAASVHVSGDFSPFGLRRDEAVRAALADLPGDVGLYATGSPYLVTPGRITKDDGNPY RVFTPFFRAWRDHGWRAPADSGPGSARWIDPSEVPGGAEIPEPPTDLELPAGETAARAQWRAFVRESLRD YSSNRNRPDLDATSRMSAHLKFGTIHPRTMAADLERDGAGDGAAAYLRELAFRDFYAAVLHHWPDSAWWN WNRTFDAIETDEGPDAQRLFDAWKAGMTGFPIVDAGMRQLAATGFVHNRVRMIVASFLVKDLHLPWQWGA RWFLDQLVDGDIANNQHGWQWTAGCGTDAAPYFRVFNPTKQGEKFDPDGVYIRRWVPELADPATVPDVHR IDGSRPADYPEPIVDHAQERAEALRRYGRLS*
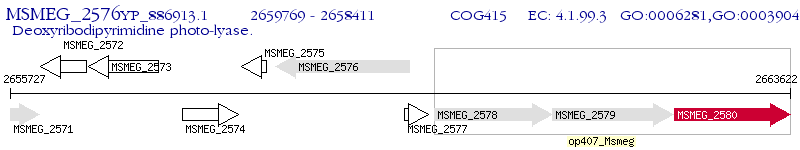
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2576 | - | - | 100% (452) | deoxyribodipyrimidine photo-lyase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4086 | - | 0.0 | 74.67% (454) | deoxyribodipyrimidine photo-lyase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1671 | phrI | 1e-176 | 67.04% (446) | DNA photolyase, PhrI |
| M. avium 104 | MAV_3897 | - | 0.0 | 69.71% (449) | Phr protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1907 | phrI | 1e-175 | 66.37% (446) | DNA photolyase, PhrI |
| M. vanbaalenii PYR-1 | Mvan_2257 | - | 0.0 | 74.33% (448) | deoxyribodipyrimidine photo-lyase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4086|M.gilvum_PYR-GCK -MPAVLWFRRDLRLADLPALVAAAEVDGDVLACYVLDPRLKATGGSRRLQ
Mvan_2257|M.vanbaalenii_PYR-1 ----MLWFRRDLRLADLPALLAASEGDGEVLACYVLDPRLKSSSGPRRLQ
MSMEG_2576|M.smegmatis_MC2_155 -MPTLLWFRRDLRCADHPAVLDAAQGDADVLACYVLDPRLTGSSGDRRLA
MMAR_1671|M.marinum_M MTRALLWFRRDLRLGDHPALAAAADSE-EVLACFVVDPRLEASSGLRRLQ
MUL_1907|M.ulcerans_Agy99 MTRALLWFRRDLRLGDHPALAAAADSE-EVLACFVVDPRLEASSGLRRLQ
MAV_3897|M.avium_104 -MPALLWFRRDLRLHDHPALSAAADSD-EVLACFVLDPRLQRSSGPRRLQ
:******** * **: *:: : :****:*:**** :.* ***
Mflv_4086|M.gilvum_PYR-GCK YLYDALRDLRESLDGRLLITRGDPVSRIPVLAAAVGASSVHVSEDFTPFG
Mvan_2257|M.vanbaalenii_PYR-1 YLYDALRDLRDNLDGRLLVTHGHPVQRIPLLAKAVGATSVHVSEDFTPFG
MSMEG_2576|M.smegmatis_MC2_155 YLYGALRDLREQLGGKLLVTRGRPEQRIPELAAAIGAASVHVSGDFSPFG
MMAR_1671|M.marinum_M FLGDSLRQLNDALQGRLLVTRGRPEQRIPLIAKAIEASSVHVSGDFAPFG
MUL_1907|M.ulcerans_Agy99 FLGDSLRQLNDALQGRLLVTRGRPEQRIPLIAKAIEASSVHVTGDFAPFG
MAV_3897|M.avium_104 FLGDSLRVLRDELDGRLLVTRGRPDIRIPEIAKAIGASSVHVSEDFTPFG
:* .:** *.: * *:**:*:* * *** :* *: *:****: **:***
Mflv_4086|M.gilvum_PYR-GCK MRRDEQVRDALG----DVELVATGSPYLVSPGRVTKGDGSPYKVFTPFFD
Mvan_2257|M.vanbaalenii_PYR-1 RRRDDQVRDALG----GVELVATGSPYLVSPGRVTKADGTPYKVFTPFFA
MSMEG_2576|M.smegmatis_MC2_155 LRRDEAVRAALADLPGDVGLYATGSPYLVTPGRITKDDGNPYRVFTPFFR
MMAR_1671|M.marinum_M RRRDERVAAGLG----RVPLVATGSPYLVSPGRVVKNDGSPYQVFTPFFR
MUL_1907|M.ulcerans_Agy99 RRRDERVAAGLG----RVPLVATGSPYLVSPGRVVKNDGSPYQVFTPFFR
MAV_3897|M.avium_104 KRRDARVRAALA----SVPLVATGSPYLVPPGRVTKPDGSPYQVFTPFLR
*** * .*. * * ********.***:.* **.**:*****:
Mflv_4086|M.gilvum_PYR-GCK AWRQHGWRPPAESGPPSAHWLDPADVAGKPGITDIPDAGVELDLPAGETA
Mvan_2257|M.vanbaalenii_PYR-1 AWRTHGWRPPAGSGPASARWIDPDDVTGGPAAADIPDTGLQLELPAGETA
MSMEG_2576|M.smegmatis_MC2_155 AWRDHGWRAPADSGPGSARWIDPSEVPGG---AEIPEPPTDLELPAGETA
MMAR_1671|M.marinum_M KWQDARWRAPAGTGPGSACWLDPAQTDIEQ--CEIPDPGTELDLAAGEAA
MUL_1907|M.ulcerans_Agy99 KWQDARWRAPADTGPGSACWLDPAQTDIEQ--CEIPDPGTELDLAAGEAA
MAV_3897|M.avium_104 RWRDTGWRPPAKTGAASARWLDPARLGITA--CEIPDPGATLDLAAGEAA
*: **.** :*. ** *:** :**:. *:*.***:*
Mflv_4086|M.gilvum_PYR-GCK ARRQWKQFVDDGLAGYAEDRNRPDLAATSRMSAHLKFGTIHPRTLAADLG
Mvan_2257|M.vanbaalenii_PYR-1 ALRAWTRFTRDGLADYDEDRNRPDLAATSRMSAHLKFGAIHPRTMAADLG
MSMEG_2576|M.smegmatis_MC2_155 ARAQWRAFVRESLRDYSSNRNRPDLDATSRMSAHLKFGTIHPRTMAADLE
MMAR_1671|M.marinum_M AIREWTAFRTAKLDRYEHDRDRPDFEGTSRMSAHLKFGTIHPRTMVAGMD
MUL_1907|M.ulcerans_Agy99 AIREWTAFRTAKLDRYEHDRDRPDFEGTSRMSAYLKFGTIHPRTMVAGMD
MAV_3897|M.avium_104 ARNQWKSFVDNGLANYADDRHRPDIEGTSRMSAHLKFGTIHPRTLVADLD
* * * * * :*.***: .******:****:*****:.*.:
Mflv_4086|M.gilvum_PYR-GCK R---GKGAQAYLRELAFRDFYAAVVAEWPRSTWWNWNSTFDAIEVDDGPD
Mvan_2257|M.vanbaalenii_PYR-1 R---GEGAQAYLRELAFRDFYASVLAEWPDSAWWNWNTAFDAIEVDDDGD
MSMEG_2576|M.smegmatis_MC2_155 RDGAGDGAAAYLRELAFRDFYAAVLHHWPDSAWWNWNRTFDAIETDEGPD
MMAR_1671|M.marinum_M LT--GTGARSYLRELAFRDFYGAVLHHWPASAWHNWNSNFDGIVTDTDAD
MUL_1907|M.ulcerans_Agy99 LT--GTGARSYLRELAFRDFYGAVLHHWPASAWHNWNSNFDGIVTDTDAN
MAV_3897|M.avium_104 LR--TAGARAYLRELAFRDFYADVLHHRPASAWRNWNSAFDAIRTDTGDE
** :***********. *: . * *:* *** **.* .* . :
Mflv_4086|M.gilvum_PYR-GCK AQRAFEAWKAGRTGFPIVDAGMRQLAETGFVHNRVRMIVASFLVKDLHLP
Mvan_2257|M.vanbaalenii_PYR-1 AQKSFAAWKQGRTGFPIVDAGMRQLAETGFMHNRVRMITASFLVKDLHLP
MSMEG_2576|M.smegmatis_MC2_155 AQRLFDAWKAGMTGFPIVDAGMRQLAATGFVHNRVRMIVASFLVKDLHLP
MMAR_1671|M.marinum_M ARRLFECWKAGETGYPLVDAGMRQLRETGFMHNRVRMIAASFLVKDLHLP
MUL_1907|M.ulcerans_Agy99 ARRVFECWKAGETGYPLVDAGMRQLRETGFMHNRVRMIAASFLVKDLHLP
MAV_3897|M.avium_104 AGRRFEAWKAGETGFPFVDAGMRQLRQTGFMHNRVRMTVASFLVKDLHLP
* : * .** * **:*:******** ***:****** .***********
Mflv_4086|M.gilvum_PYR-GCK WQWGARWFLDQLTDGDMANNQHGWQWAAGCGTDAAPYFRVFNPTTQGKKF
Mvan_2257|M.vanbaalenii_PYR-1 WQWGARWFLEQLVDGDIANNQHGWQWAAGCGTDAAPYFRVFNPSTQGKKF
MSMEG_2576|M.smegmatis_MC2_155 WQWGARWFLDQLVDGDIANNQHGWQWTAGCGTDAAPYFRVFNPTKQGEKF
MMAR_1671|M.marinum_M WQWGAAWFLDQLTDGDLASNQHGWQWSAGCGTDAAPYFRIFNPSAQAEKF
MUL_1907|M.ulcerans_Agy99 WQWGAAWFLDQLTDGDLASNQHGWQWSAGCGTDAAPYFRIFNPSAQAEKF
MAV_3897|M.avium_104 WQWGAHWFLQQLVDGDLANNQHGWQWCAGCGTDAAPYFRVFNPAAQGEKF
***** ***:**.***:*.******* ************:***: *.:**
Mflv_4086|M.gilvum_PYR-GCK DPAGVYVKRWVEELRDVASSVDVHNLDRDRPADYPEPIVDHAQERAEALR
Mvan_2257|M.vanbaalenii_PYR-1 DPSGDYVRRWVPELADVD---DVHKLDGDRPDGYPEPIVDHGQERAEALR
MSMEG_2576|M.smegmatis_MC2_155 DPDGVYIRRWVPELADPATVPDVHRIDGSRPADYPEPIVDHAQERAEALR
MMAR_1671|M.marinum_M DPAGDYIRRWVPELRDVDNVHRIQRRGGPCPSTYPAPIVDHGTERAEALR
MUL_1907|M.ulcerans_Agy99 DPAGDYIRRWVPELRDVDNVHRIQRRGGPCPSTYPAPIVDHGTERAEALR
MAV_3897|M.avium_104 DPSGDYIRRWVPELRCADDPH---LRTGERPPGYPAPIVDHATERAEALR
** * *::*** ** * ** *****. *******
Mflv_4086|M.gilvum_PYR-GCK RYNSLRG
Mvan_2257|M.vanbaalenii_PYR-1 RYRAIG-
MSMEG_2576|M.smegmatis_MC2_155 RYGRLS-
MMAR_1671|M.marinum_M RYHDIG-
MUL_1907|M.ulcerans_Agy99 RYHDIG-
MAV_3897|M.avium_104 RYRSM--
** :