For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPSDNRQPEEVWLAMAVVTMKQLLDSGAHFGHQTRRWNPKMKRFIFTDRNGIYIIDLQQTLTYIDKAYEF VKETVAHGGTVLFVGTKKQAQESIAEEATRVGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGF EGRTKKEILMLTREKNKLERSLGGIRDMQKVPSAVWVVDTNKEHIAVGEARKLGIPVIAILDTNCDPDVV DYPIPGNDDAIRSAALLTKVIASAVAEGLQARAGQGSGEKPAEGAEPLAEWEQELLAGATAGAADASAEG AAAPESSTDAS
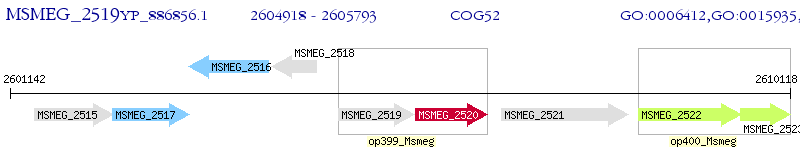
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2519 | rpsB | - | 100% (291) | 30S ribosomal protein S2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2914c | rpsB | 1e-137 | 86.41% (287) | 30S ribosomal protein S2 |
| M. gilvum PYR-GCK | Mflv_4133 | rpsB | 1e-138 | 87.81% (279) | 30S ribosomal protein S2 |
| M. tuberculosis H37Rv | Rv2890c | rpsB | 1e-137 | 86.41% (287) | 30S ribosomal protein S2 |
| M. leprae Br4923 | MLBr_01598 | rpsB | 1e-131 | 88.30% (265) | 30S ribosomal protein S2 |
| M. abscessus ATCC 19977 | MAB_3196c | rpsB | 1e-138 | 87.64% (275) | 30s ribosomal protein S2 |
| M. marinum M | MMAR_1819 | rpsB | 1e-138 | 91.27% (275) | 30S ribosomal protein S2 RpsB |
| M. avium 104 | MAV_3744 | rpsB | 1e-139 | 89.96% (279) | 30S ribosomal protein S2 |
| M. thermoresistible (build 8) | TH_0150 | rpsB | 1e-139 | 91.85% (270) | PROBABLE 30S RIBOSOMAL PROTEIN S2 RPSB |
| M. ulcerans Agy99 | MUL_2067 | rpsB | 1e-135 | 90.74% (270) | 30S ribosomal protein S2 |
| M. vanbaalenii PYR-1 | Mvan_2209 | rpsB | 1e-142 | 90.84% (273) | 30S ribosomal protein S2 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2519|M.smegmatis_MC2_155 MPSDNRQPEEVWLAMAVVTMKQLLDSGAHFGHQTRRWNPKMKRFIFTDRN
TH_0150|M.thermoresistible__bu -------------------MKQLLDAGAHFGHQTRRWNPKMKRFIFTDRN
MMAR_1819|M.marinum_M --------------MAVVTMKQLLDSGTHFGHQTRRWNPKMKRFIFTDRN
MUL_2067|M.ulcerans_Agy99 -------------------MKQLLDSGTHFGHQTRRWNPKMKRFIFTDRN
Mb2914c|M.bovis_AF2122/97 --------------MAVVTMKQLLDSGTHFGHQTRRWNPKMKRFIFTDRN
Rv2890c|M.tuberculosis_H37Rv --------------MAVVTMKQLLDSGTHFGHQTRRWNPKMKRFIFTDRN
MAV_3744|M.avium_104 --------------MAVVTMKQLLDSGTHFGHQTRRWNPKMKRFIFTDRN
MLBr_01598|M.leprae_Br4923 --------------MAVVTMKQLLDSGTHFGHQTRRWNPKMKRFIFTDRN
Mflv_4133|M.gilvum_PYR-GCK --------------MAVVTMKQLLDSGAHFGHQTRRWNPKMKRFIFTDRN
Mvan_2209|M.vanbaalenii_PYR-1 --------------MAVVTMKQLLDSGAHFGHQTRRWNPKMKRFIFTDRN
MAB_3196c|M.abscessus_ATCC_199 --------------MAVVTMKQLLDSGAHFGHQTRRWNPKMKRFIFTDRN
******:*:**********************
MSMEG_2519|M.smegmatis_MC2_155 GIYIIDLQQTLTYIDKAYEFVKETVAHGGTVLFVGTKKQAQESIAEEATR
TH_0150|M.thermoresistible__bu GIYIIDLQQTLSYIDKAYEFVKETVAHGGTILFVGTKKQAQEAIAEEATR
MMAR_1819|M.marinum_M GIYIIDLQQTLTFIDKAYEFVKETVAHGGTVLFVGTKKQAQESVAAEATR
MUL_2067|M.ulcerans_Agy99 GIYIIDLQQTLTFIDKAYEFVKETVAHGGTVLFVGTKKQAQESVAAEATR
Mb2914c|M.bovis_AF2122/97 GIYIIDLQQTLTFIDKAYEFVKETVAHGGSVLFVGTKKQAQESVAAEATR
Rv2890c|M.tuberculosis_H37Rv GIYIIDLQQTLTFIDKAYEFVKETVAHGGSVLFVGTKKQAQESVAAEATR
MAV_3744|M.avium_104 GIYIIDLQQTLTFIDKAYEFVKETVAHGGTVLFVGTKKQAQESIAAEATR
MLBr_01598|M.leprae_Br4923 GIYIIDLQQTLTFIDKAYEFVKETVARGGLVLFVGTKKQAQESVAAEATR
Mflv_4133|M.gilvum_PYR-GCK GIYIIDLQQTLTYIDSAYEFVKETVAHGGSIMFVGTKKQAQESIAQEATR
Mvan_2209|M.vanbaalenii_PYR-1 GIYIIDLQQTLTYIDKAYEFVKETVAHGGSIMFVGTKKQAQESIAEEATR
MAB_3196c|M.abscessus_ATCC_199 GIYIIDLQQTLTYIDKAYEFVKETVAHGGSVMFVGTKKQAQEPIAEEATR
***********::**.**********:** ::**********.:* ****
MSMEG_2519|M.smegmatis_MC2_155 VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGFEGRTKKEILM
TH_0150|M.thermoresistible__bu VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGFEGRTKKEILM
MMAR_1819|M.marinum_M VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGFEGRTKKEILG
MUL_2067|M.ulcerans_Agy99 VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGFEGRSKKEILG
Mb2914c|M.bovis_AF2122/97 VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGFEGRTKKEILG
Rv2890c|M.tuberculosis_H37Rv VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGFEGRTKKEILG
MAV_3744|M.avium_104 VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGFEGRTKKEILG
MLBr_01598|M.leprae_Br4923 VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGFEGRTKKEILG
Mflv_4133|M.gilvum_PYR-GCK VGMPYVNQRWLGGMLTNFQTVHKRLQRMKELEAMEQTGGFEGRTKKEILM
Mvan_2209|M.vanbaalenii_PYR-1 VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELEAMEQTGGFEGRTKKEILM
MAB_3196c|M.abscessus_ATCC_199 VGMPYVNQRWLGGMLTNFSTVHKRLQRLKELESMEQTGGFEGRTKKEILM
******************.********:****:**********:*****
MSMEG_2519|M.smegmatis_MC2_155 LTREKNKLERSLGGIRDMQKVPSAVWVVDTNKEHIAVGEARKLGIPVIAI
TH_0150|M.thermoresistible__bu LTREKNKLERSLGGIRDMQKVPSAIWVVDTNKEHIAVGEARKLGIPVIAI
MMAR_1819|M.marinum_M LTREKNKLERSLGGIRDMAKVPSAIWVVDTNKEHIAVGEARKLGIPVIAI
MUL_2067|M.ulcerans_Agy99 LTREKNKLERSLGGIRDMAKVPSAIWVVDTNKEHIAVGEARKLGIPVIAI
Mb2914c|M.bovis_AF2122/97 LTREKNKLERSLGGIRDMAKVPSAIWVVDTNKEHIAVGEARKLGIPVIAI
Rv2890c|M.tuberculosis_H37Rv LTREKNKLERSLGGIRDMAKVPSAIWVVDTNKEHIAVGEARKLGIPVIAI
MAV_3744|M.avium_104 LTREKNKLERSLGGIRDMSKVPSAIWVVDTNKEHIAVGEARKLGIPVIAI
MLBr_01598|M.leprae_Br4923 LTREKNKLERSLGGIRDMAKVPSAIWVVDTNKEHIAVGEARKLGVPVIAI
Mflv_4133|M.gilvum_PYR-GCK LTREKNKLERSLGGIRDMAKVPSAIWVVDTNKEHLAVAEARKLNIPIIAI
Mvan_2209|M.vanbaalenii_PYR-1 LTREKNKLERSLGGIRDMQKVPSAIWVVDTNKEHLAVAEARKLNIPIIAI
MAB_3196c|M.abscessus_ATCC_199 LTREKNKLERSLGGIRDMQKVPSAIFVVDTNKEHLAVAEARKLNIPIIAI
****************** *****::********:**.*****.:*:***
MSMEG_2519|M.smegmatis_MC2_155 LDTNCDPDVVDYPIPGNDDAIRSAALLTKVIASAVAEGLQARAGQG-SGE
TH_0150|M.thermoresistible__bu LDTNCDPDLVDYPIPGNDDAIRSAALLTKVIASAVAEGLQARAGAGREGD
MMAR_1819|M.marinum_M LDTNCDPDEVDYPIPGNDDAIRSAALLTKVIASAVAEGLQARAGAGRGDG
MUL_2067|M.ulcerans_Agy99 LDTNCDPDEVDYPIPGNDDAIRSAALLTKVIASAVAEGLQARAGAGRGDG
Mb2914c|M.bovis_AF2122/97 LDTNCDPDEVDYPIPGNDDAIRSAALLTRVIASAVAEGLQARAGLGRADG
Rv2890c|M.tuberculosis_H37Rv LDTNCDPDEVDYPIPGNDDAIRSAALLTRVIASAVAEGLQARAGLGRADG
MAV_3744|M.avium_104 LDTNCDPDEVDYPIPGNDDAIRSAALLTKVIASAVAEGLQARAGVGRGDG
MLBr_01598|M.leprae_Br4923 LDTNCDPDEVDYPIPGNDDAIRSAELLTTVIASAVAEGLQVRAGLRCGDG
Mflv_4133|M.gilvum_PYR-GCK LDTNCDPDVVDYPIPGNDDAIRSAALLTKVIASAVAEGLQARAGAGADKA
Mvan_2209|M.vanbaalenii_PYR-1 LDTNCDPDLVDYPIPGNDDAIRSAALLTKVVASAVAEGLQARAGAGADKA
MAB_3196c|M.abscessus_ATCC_199 LDTNCDPDVVDYPIPGNDDAIRSAALLTKVVASAIAEGLQARSGLAGGDE
******** *************** *** *:***:*****.*:*
MSMEG_2519|M.smegmatis_MC2_155 KP---AEGAEPLAEWEQELLAGATAGAADAS--------AEGAAAPESST
TH_0150|M.thermoresistible__bu KP---AETAEPLPEWEQELLAGATATAAGN---------PEGEKPAESTT
MMAR_1819|M.marinum_M KPV--VEAAEPLAEWEQELLAGATAAAPAEG-----------AVATETTP
MUL_2067|M.ulcerans_Agy99 KPV--VEAAEPLAEWEQELLAGATAAAPAEG-----------AVATETTP
Mb2914c|M.bovis_AF2122/97 KPE--AEAAEPLAEWEQELLASATASATPSATASTTALTDAPAGATEPTT
Rv2890c|M.tuberculosis_H37Rv KPE--AEAAEPLAEWEQELLASATASATPSATASTTALTDAPAGATEPTT
MAV_3744|M.avium_104 KPE--VEAAEPLAEWEQELLASATATAAP-----------TEAGAPEPTT
MLBr_01598|M.leprae_Br4923 KPE--TEAVEPLPEWEQELLAGAGSSALNDS----------GADLSEANP
Mflv_4133|M.gilvum_PYR-GCK EAGQDGAAAEPLAEWEQELLAGATTAAPEAA--------AGEAAAAPEQS
Mvan_2209|M.vanbaalenii_PYR-1 AA----EAAEPLAEWEQELLAGATTAAPEAA--------AGEAAAP-EQS
MAB_3196c|M.abscessus_ATCC_199 KP----EAGEPLAEWEQELLASAVATTDEAS--------APSAAATETTT
. ***.********.* : : . .
MSMEG_2519|M.smegmatis_MC2_155 DAS-
TH_0150|M.thermoresistible__bu PQNS
MMAR_1819|M.marinum_M TEG-
MUL_2067|M.ulcerans_Agy99 TEG-
Mb2914c|M.bovis_AF2122/97 DAS-
Rv2890c|M.tuberculosis_H37Rv DAS-
MAV_3744|M.avium_104 DPS-
MLBr_01598|M.leprae_Br4923 TEA-
Mflv_4133|M.gilvum_PYR-GCK S---
Mvan_2209|M.vanbaalenii_PYR-1 ----
MAB_3196c|M.abscessus_ATCC_199 EEG-