For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDLWVERTGVRRYTGRSSRGAEVLVGNEDVEGVFTPGELMKIALAACTGMASDQPLSRRLGEDYPATIR VSGAADREREVYPHLEEKLEIDLSGLSEADVKRVLTVVERAVDQVCTVGRTLKSGTEVTFSVADTVLPK
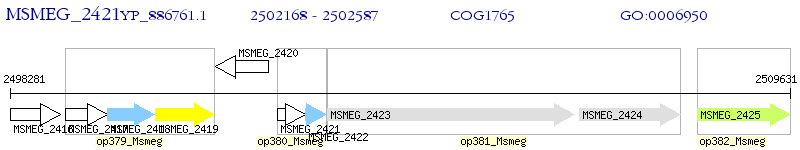
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2421 | - | - | 100% (139) | hypothetical protein MSMEG_2421 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2948c | - | 4e-52 | 70.15% (134) | hypothetical protein Mb2948c |
| M. gilvum PYR-GCK | Mflv_4190 | - | 7e-59 | 80.60% (134) | OsmC family protein |
| M. tuberculosis H37Rv | Rv2923c | - | 4e-52 | 70.15% (134) | hypothetical protein Rv2923c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3248c | - | 2e-41 | 73.39% (109) | hypothetical protein MAB_3248c |
| M. marinum M | MMAR_1784 | - | 5e-55 | 75.37% (134) | hypothetical protein MMAR_1784 |
| M. avium 104 | MAV_3780 | - | 8e-43 | 75.45% (110) | hypothetical protein MAV_3780 |
| M. thermoresistible (build 8) | TH_2279 | - | 1e-60 | 82.84% (134) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2032 | - | 4e-55 | 75.37% (134) | hypothetical protein MUL_2032 |
| M. vanbaalenii PYR-1 | Mvan_2173 | - | 9e-60 | 83.58% (134) | OsmC family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4190|M.gilvum_PYR-GCK MHSSRSPLDLRLLECNFGGGCRNRGVTELWVERTGVRRYTGRSTRGAEVL
Mvan_2173|M.vanbaalenii_PYR-1 -------------------------MTELWVERTGVRRYTGRSTRGAEVL
MSMEG_2421|M.smegmatis_MC2_155 -------------------------MTDLWVERTGVRRYTGRSSRGAEVL
TH_2279|M.thermoresistible__bu -------------------------MTELWVERTGARRYTGRSSRGAEVL
MAB_3248c|M.abscessus_ATCC_199 ------------------------------------------------ML
MAV_3780|M.avium_104 ------------------------------------------------ML
Mb2948c|M.bovis_AF2122/97 -------------------------MTQLWVERTGTRRYIGRSTRGAQVL
Rv2923c|M.tuberculosis_H37Rv -------------------------MTQLWVERTGTRRYIGRSTRGAQVL
MMAR_1784|M.marinum_M -------------------------MTQLWVERTGARRYTGHSSRGAQVL
MUL_2032|M.ulcerans_Agy99 -------------------------MTQLWVERTGARRYTGHSSRGAQVL
:*
Mflv_4190|M.gilvum_PYR-GCK VGSEDVEGVFTPGELMKIALAACSGLSSDQPLRRRLGDDYRAKIRVSGAA
Mvan_2173|M.vanbaalenii_PYR-1 VGSEDVEGVFTPGELMKIALAACSGMSSDQPLRRRLGDDYQATIRVSGPA
MSMEG_2421|M.smegmatis_MC2_155 VGNEDVEGVFTPGELMKIALAACTGMASDQPLSRRLGEDYPATIRVSGAA
TH_2279|M.thermoresistible__bu IGSEDVEGVFTPGELLKIALAACSGMASDQPLRRRLGDDYPATIRVSGAA
MAB_3248c|M.abscessus_ATCC_199 IGSETVDGVFTPGELLKIALAACSGMSSDQPLARRLGEDYRVRIEVSGPA
MAV_3780|M.avium_104 IGSEDVDGVFTPGELLKIALAACSGMASDAPLARRLGDDYRAVIRVSGAA
Mb2948c|M.bovis_AF2122/97 VGSEDVDGVFTPGELLKIALAACSGMASDQPLARRLGDDYQAVVKVSGAA
Rv2923c|M.tuberculosis_H37Rv VGSEDVDGVFTPGELLKIALAACSGMASDQPLARRLGDDYQAVVKVSGAA
MMAR_1784|M.marinum_M IGSEDVDGVFTPGELMKIALAACSGMSSDRPLARRLGDDYQAVVRVSGAA
MUL_2032|M.ulcerans_Agy99 IGSEDVDGVFTPGELMKIALAACSGMSSDRPLARRLGDDYQAVVRVSGAA
:*.* *:********:*******:*::** ** ****:** . :.***.*
Mflv_4190|M.gilvum_PYR-GCK DREQERYPLLEEKLEVDLSGLSDDEKARLVTVVERAIDQVCTVGRTLKAG
Mvan_2173|M.vanbaalenii_PYR-1 DREQERYPLLEEVLEIDLSGLSDEDRDRVLTVVERAIDQVCTVGRTLKSG
MSMEG_2421|M.smegmatis_MC2_155 DREREVYPHLEEKLEIDLSGLSEADVKRVLTVVERAVDQVCTVGRTLKSG
TH_2279|M.thermoresistible__bu DREQERYPLLEEKLELDLSGLSEQERDRVLTVVQRAIDQVCTVGRTLQSG
MAB_3248c|M.abscessus_ATCC_199 DREQERYPVLDEKMVIDLSGLTSEERQRVLTVVERAVDQVCTVGRTLKAG
MAV_3780|M.avium_104 DREQERYPLLEETLELDLSGLSDEDRQRLLLVVHRAVDQVCTVGRTLTSG
Mb2948c|M.bovis_AF2122/97 DRDQERYPLIEETMELDLSGLTEDEKERLLVVINRAVELACTVGRTLKSG
Rv2923c|M.tuberculosis_H37Rv DRDQERYPLIEETMELDLSGLTEDEKERLLVVINRAVELACTVGRTLKSG
MMAR_1784|M.marinum_M DRDQERYPLLEETLELDLSGLSEEEKERVRVVVDRAIDLVCTVGRTLKSG
MUL_2032|M.ulcerans_Agy99 DRDQERYPLLEETLELDLSGLSEEEKERVRVVVDRAIDLVCTVGRTLKSG
**::* ** ::* : :*****:. : *: *:.**:: .******* :*
Mflv_4190|M.gilvum_PYR-GCK TEVTF---EVGDAGGE----------------------------------
Mvan_2173|M.vanbaalenii_PYR-1 TEVTFGVTEVKNVGGK----------------------------------
MSMEG_2421|M.smegmatis_MC2_155 TEVTFS--VADTVLPK----------------------------------
TH_2279|M.thermoresistible__bu TKVDF---EVVDVGRD----------------------------------
MAB_3248c|M.abscessus_ATCC_199 AEVNFSIEP-----------------------------------------
MAV_3780|M.avium_104 TTVNFEIAAGAGHAGA----------------------------------
Mb2948c|M.bovis_AF2122/97 TTVNLEVVDVGA--------------------------------------
Rv2923c|M.tuberculosis_H37Rv TTVNLEVVDVGA--------------------------------------
MMAR_1784|M.marinum_M TVVNFEVADVAPVSQPDVRLTAWVHGHVQGVGFRWWTRCRALELGLTGYA
MUL_2032|M.ulcerans_Agy99 TVVNFEVADVAPVSQPDVRLTAWVHGHVQGVGFRWWTRCRALELGLTGYA
: * :
Mflv_4190|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2173|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2421|M.smegmatis_MC2_155 --------------------------------------------------
TH_2279|M.thermoresistible__bu --------------------------------------------------
MAB_3248c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_3780|M.avium_104 --------------------------------------------------
Mb2948c|M.bovis_AF2122/97 --------------------------------------------------
Rv2923c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1784|M.marinum_M ANHADGRVMVVAQGPRDSGVQLLRLLEGDTTPGRVDKVVADWSQRVEPIS
MUL_2032|M.ulcerans_Agy99 ANHADGRVMVVAQGPRDSGVQLLRLLEGDTTPGRVDKVVADWSQRVEPIS
Mflv_4190|M.gilvum_PYR-GCK -----
Mvan_2173|M.vanbaalenii_PYR-1 -----
MSMEG_2421|M.smegmatis_MC2_155 -----
TH_2279|M.thermoresistible__bu -----
MAB_3248c|M.abscessus_ATCC_199 -----
MAV_3780|M.avium_104 -----
Mb2948c|M.bovis_AF2122/97 -----
Rv2923c|M.tuberculosis_H37Rv -----
MMAR_1784|M.marinum_M GFSER
MUL_2032|M.ulcerans_Agy99 GFSER