For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VADSKPKTFVQSTDDVVRFLTDQHNLIKDLFEEVLSASGDDARQTAFVELRQLLAVHETAEEMVVHPRAR AAVDGGDKIVDARLEEEHKAKQQLSELEKLDIGSAEFITELEKFREAVLDHAAHEEAEEFVKLSRELDAQ ELERMTKAVQAAEAIAPTRPHAGVESASLNFAVGPFASMLDRARDLIGSALK
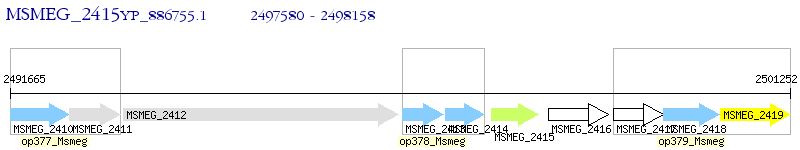
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2415 | - | - | 100% (192) | hemerythrin HHE cation binding region |
| M. smegmatis MC2 155 | MSMEG_3312 | - | 2e-16 | 28.09% (178) | hemerythrin HHE cation binding domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_6212 | - | 1e-15 | 28.81% (177) | hemerythrin HHE cation binding domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4195 | - | 5e-71 | 71.66% (187) | hemerythrin HHE cation binding domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2504c | - | 1e-27 | 39.57% (187) | hypothetical protein MAB_2504c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3982 | - | 4e-61 | 65.38% (182) | hemerythrin HHE cation binding domain-containing protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2168 | - | 2e-74 | 73.80% (187) | hemerythrin HHE cation binding domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4195|M.gilvum_PYR-GCK ----MPETFVQSTDDVVAFLKDQHNLIRDMFDDVIHASDPKAREKAFTEL
Mvan_2168|M.vanbaalenii_PYR-1 ----MPETFVQSTDDVVAFLKDQHKLIKDMFDDVIHASEPKAREKAFTDL
MAV_3982|M.avium_104 ----MADMIVKSPDEVVAFLKAQHNLIEDMFDQVLHATDPKAREEPFATL
MSMEG_2415|M.smegmatis_MC2_155 MADSKPKTFVQSTDDVVRFLTDQHNLIKDLFEEVLSASGDDARQTAFVEL
MAB_2504c|M.abscessus_ATCC_199 -MASLPTPELATASDLVEFLEDQHVAIRTLFDAALSP-EKGIDSGSFTRL
. : :..::* ** ** *. :*: .: . . .*. *
Mflv_4195|M.gilvum_PYR-GCK RQLLAVHETAEEMVVHPRIRREVDEGDDIAEALLAEEHASKEKLVALENL
Mvan_2168|M.vanbaalenii_PYR-1 RQLLAVHETAEEMVVHPRARREIAAGDQVVDARLKEEHAAKEQLVALENL
MAV_3982|M.avium_104 RQLLAVHETAEEMLVHPRARKEADAGDAVVDARLHEEHSAKELLSAIEKL
MSMEG_2415|M.smegmatis_MC2_155 RQLLAVHETAEEMVVHPRARAAVDGGDKIVDARLEEEHKAKQQLSELEKL
MAB_2504c|M.abscessus_ATCC_199 RGLLAVHETVEAMVVHPWSRGISEAGRTVANTRLAEELALTTDLEQLEAT
* *******.* *:*** * * :.:: * ** . * :*
Mflv_4195|M.gilvum_PYR-GCK DIASDEFLRELTEFRDTVLAHAEHEEIEEFTKLSRELGADDLKRMAGAVQ
Mvan_2168|M.vanbaalenii_PYR-1 DIASDEFITELTKFRDAVLEHAEHEEAEEFTKLKRELDADELKSLAGAVR
MAV_3982|M.avium_104 DSTTDQFLDEVTKLREAVLEHATREENEEFPALQR-LDSDDLKRMGTAVR
MSMEG_2415|M.smegmatis_MC2_155 DIGSAEFITELEKFREAVLDHAAHEEAEEFVKLSRELDAQELERMTKAVQ
MAB_2504c|M.abscessus_ATCC_199 AVGSPEFIVGLAALRSATLEHCRLEEEEEFPMLSGELGPVDRRRAGLAVG
: :*: : :*.:.* *. ** *** *. *.. : . **
Mflv_4195|M.gilvum_PYR-GCK AAEAIAPTRPHPGVESATLNFAVGPFASMLDRARDVIGAALK
Mvan_2168|M.vanbaalenii_PYR-1 AAEAIAPTRPHPGVESATLNFAVGPFASMLDRARDAIGAALR
MAV_3982|M.avium_104 AAEAIAPTHPHPGVESATLNFAVGPFASMLDRARDLIGRAIG
MSMEG_2415|M.smegmatis_MC2_155 AAEAIAPTRPHAGVESASLNFAVGPFASMLDRARDLIGSALK
MAB_2504c|M.abscessus_ATCC_199 MSEWFAPAPERSPAESTIASPTHGPYGEMAARVRDILAAV--
:* :**: :. .**: . : **:..* *.** :. .