For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLVEHADGALKKVSAELLTAARALGEPSAVVVGKPGTAAGLTDGLKAAGAAKIYVAESDDVENYLLTPYV DVLAALAESAAPAGVIIAANADGKEIAGRLAARIGSGLLVDVIEVKEGGVGVHSIFGGAFTVDAQVTSET PVITVRPGSVEAVPADGAGEVVNVEVPAQAENATRITKREPAVAGDRPELTEASVVVAGGRGVGSAENFK IVEELADSLGGAVGASRAAVDSGYYPGQFQVGQTGKTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKD EEAPIFEIADLGIVGDLFKVSPQLTEAVKARKG
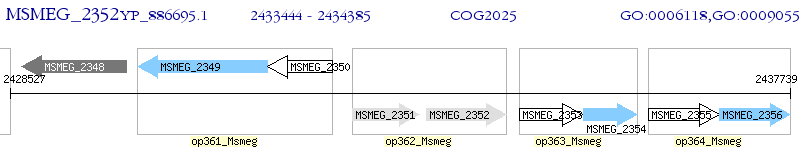
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2352 | etfA | - | 100% (313) | electron transfer flavoprotein, alpha subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3054c | fixB | 1e-149 | 86.26% (313) | electron transfer flavoprotein subunit alpha |
| M. gilvum PYR-GCK | Mflv_4258 | - | 1e-154 | 87.54% (313) | electron transfer flavoprotein, alpha subunit |
| M. tuberculosis H37Rv | Rv3028c | fixB | 1e-149 | 86.58% (313) | electron transfer flavoprotein subunit alpha |
| M. leprae Br4923 | MLBr_01711 | eftA | 1e-141 | 80.19% (313) | electron transfer flavoprotein alpha subunit |
| M. abscessus ATCC 19977 | MAB_3362c | - | 1e-141 | 80.19% (313) | electron transfer flavoprotein subunit alpha FixB |
| M. marinum M | MMAR_1685 | fixB | 1e-147 | 84.03% (313) | electron transfer flavoprotein (alpha-subunit) FixB |
| M. avium 104 | MAV_3875 | - | 1e-147 | 85.21% (311) | electron transfer flavoprotein, alpha subunit |
| M. thermoresistible (build 8) | TH_2195 | fixB | 1e-148 | 84.03% (313) | PROBABLE ELECTRON TRANSFER FLAVOPROTEIN (ALPHA-SUBUNIT) FIXB |
| M. ulcerans Agy99 | MUL_1923 | fixB | 1e-148 | 84.03% (313) | electron transfer flavoprotein (alpha-subunit) FixB |
| M. vanbaalenii PYR-1 | Mvan_2100 | - | 1e-152 | 86.26% (313) | electron transfer flavoprotein, alpha subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1685|M.marinum_M MAEVLVLVEHAEGALKKVSAELITAARALGEPAAVVIGAPGTAAPLVDGL
MUL_1923|M.ulcerans_Agy99 MAEVLVLVEHAEGALKKVSAELITAARALGEPAAVVIGAPGTAAPLVDGL
Mb3054c|M.bovis_AF2122/97 MAEVLVLVEHAEGALKKVSAELITAARALGEPAAVVVGVPGTAAPLVDGL
Rv3028c|M.tuberculosis_H37Rv MAEVLVLVEHAEGALKKVSAELITAARALGEPAAVVVGVPGTAAPLVDGL
MLBr_01711|M.leprae_Br4923 MAEALVLVEHTEGALKKVSAELITAARVLGEPAAVVVGTPGTSAPLVDGL
MAB_3362c|M.abscessus_ATCC_199 MAEVLVLVEHSEGALKKVSAELITAARVLGEPSAVVAGPAGTAAPLIDGL
MAV_3875|M.avium_104 -------MEHAEGALKKVTSELITAARALGEPSAVVVGTPGTAAPLVDGL
Mflv_4258|M.gilvum_PYR-GCK MAEVLVLVEHAEGALKKVTAELITAAKKLGEPSAVVVGKPGTSEGLVDGL
Mvan_2100|M.vanbaalenii_PYR-1 MAEVLVLVEHAEGALKKVTSELITAARVLGEPAAVVVGKPGTAEGLIDGL
MSMEG_2352|M.smegmatis_MC2_155 -----MLVEHADGALKKVSAELLTAARALGEPSAVVVGKPGTAAGLTDGL
TH_2195|M.thermoresistible__bu -----VLVEHAEGALKKVTTELITAARRLGEPSAVVVGAPGTAEPLIEGL
:**::******::**:***: ****:*** * .**: * :**
MMAR_1685|M.marinum_M KAAGAAKIYVAESDVVDQYLITPFVDVLAGLAESSAPAAVLVAATADGKE
MUL_1923|M.ulcerans_Agy99 KAAGAAKIYVAESDVVDQYLITPFVDVLAGLAESSAPAAVLVAATADGKE
Mb3054c|M.bovis_AF2122/97 KAAGAAKIYVAESDLVDKYLITPAVDVLAGLAESSAPAGVLIAATADGKE
Rv3028c|M.tuberculosis_H37Rv KAAGAAKIYVAESDLVDKYLITPAVDVLAGLAESSAPAGVLIAATADGKE
MLBr_01711|M.leprae_Br4923 KTAGAAKIYVAESDAADKYLITPVVDVLAALAESSAPAAVLLAATADGKE
MAB_3362c|M.abscessus_ATCC_199 KEAGAAKIYVAESDVVDSYLVTPKVDILAALAETASPAGILIAATTEGKE
MAV_3875|M.avium_104 KEAGAAKIYVAESDDVDKYLITPVVDVLASLAESNAPAAVLLAANADGKE
Mflv_4258|M.gilvum_PYR-GCK KSAGAAKIYVAESDDAENYLITPYVDVLAALAESATSAGVVLAASADGKE
Mvan_2100|M.vanbaalenii_PYR-1 KSAGAVKIYVAESDDAESYLITPYVDVLASLAESATPAAVLLASSADGKE
MSMEG_2352|M.smegmatis_MC2_155 KAAGAAKIYVAESDDVENYLLTPYVDVLAALAESAAPAGVIIAANADGKE
TH_2195|M.thermoresistible__bu KAAGAAKIYVAESDDVPNYLITPQLDVLAGLVESASPAAVLLAASADGKE
* ***.******** . .**:** :*:**.*.*: :.*.:::*:.::***
MMAR_1685|M.marinum_M IAGRLAARIGSGLLVDVVGINDGGKAVHSIFGGAFTVEAQATGDTPVISV
MUL_1923|M.ulcerans_Agy99 IAGRLAARIGSGLLVDVVGINDGGKAVHSIFGGAFTVEAQATGDTPVISV
Mb3054c|M.bovis_AF2122/97 IAGRLAARIGSGLLVDVVDVREGGVGVHSIFGGAFTVEAQANGDTPVITV
Rv3028c|M.tuberculosis_H37Rv IAGRLAARIGSGLLVDVVDVREGGVGVHSIFGGAFTVEAQANGDTPVITV
MLBr_01711|M.leprae_Br4923 IGGRLAARIGSGLLVDVVDVREGAVGVHSVFGGVFIVEAQANGDTPVITV
MAB_3362c|M.abscessus_ATCC_199 VAGRLAARLGSGLLTDVVEVREGGKALHSIFGGAFTVEAQANGDAPVITV
MAV_3875|M.avium_104 IAGRLAARIGSGLLVDVVEVKEGAKAVHSIFGGAFTVEAQANGDTPVITL
Mflv_4258|M.gilvum_PYR-GCK IAGRLAARIGSGILSDVVEVKEGGAGVHSIFGGAYTVEAEAVGDTPVITV
Mvan_2100|M.vanbaalenii_PYR-1 IAGRLAARIGSGILSDVVEVRDGGVGIHSIFGGAFTVEAEAVGDTPVITV
MSMEG_2352|M.smegmatis_MC2_155 IAGRLAARIGSGLLVDVIEVKEGGVGVHSIFGGAFTVDAQVTSETPVITV
TH_2195|M.thermoresistible__bu IAGRLAARTGAGILTDVVDVQPGGKGVHSIFGGAFTVEAEVTGELAIYTV
:.****** *:*:* **: :. *. .:**:***.: *:*:. .: .: ::
MMAR_1685|M.marinum_M RAGAIEAEPADGAGEQVSVDVPAPAENATKITSREPAVAGDRPELTEATV
MUL_1923|M.ulcerans_Agy99 RAGAIEAEPADGAGEQVSVDVPAPAENATKITSREPAVAGDRPELTEATV
Mb3054c|M.bovis_AF2122/97 RAGAVEAEPAAGAGEQVSVEVPAAAENAARITAREPAVAGDRPELTEATI
Rv3028c|M.tuberculosis_H37Rv RAGAVEAEPAAGAGEQVSVEVPAAAENAARITAREPAVAGDRPELTEATI
MLBr_01711|M.leprae_Br4923 RAGAVEAQPAEGAGEQVSVEVPAPAENATKITARAPAVVDNRPDLTEATV
MAB_3362c|M.abscessus_ATCC_199 RPGAVEAAPAAGAGEQVAVEVPAPAENATKVTSRQPVVGGDRPELTEASI
MAV_3875|M.avium_104 RAGAVDAEPAAGAGEQVTVEVPAAAENATKITAREPAVAGDRPELTEASI
Mflv_4258|M.gilvum_PYR-GCK RPGAIEAEPSDGAGEVVNVEVPAPAENATKITSREPAVAGDRPELTEASV
Mvan_2100|M.vanbaalenii_PYR-1 RPGAVEAAPAAGAGEVVNVEVPAPAENATKITSREPAVAGDRPELTEATV
MSMEG_2352|M.smegmatis_MC2_155 RPGSVEAVPADGAGEVVNVEVPAQAENATRITKREPAVAGDRPELTEASV
TH_2195|M.thermoresistible__bu RPGAIEAEPAEGAGEVVNVEVPAPAENATKITAREPAVAGDRPELTEASV
*.*:::* *: **** * *:*** ****:::* * *.* .:**:****::
MMAR_1685|M.marinum_M VVSGGRGVGSAENFSVVEALADSLGGAVGASRAAVDSGYYPGQFQVGQTG
MUL_1923|M.ulcerans_Agy99 VVSGGRGVGSAENFSVVEALADSLGGAVGASRAAVDSGYYPGQFQVGQTG
Mb3054c|M.bovis_AF2122/97 VVAGGRGVGSAENFSVVEALADSLGAAVGASRAAVDSGYYPGQFQVGQTG
Rv3028c|M.tuberculosis_H37Rv VVAGGRGVGSAENFSVVEALADSLGAAVGASRAAVDSGYYPGQFQVGQTG
MLBr_01711|M.leprae_Br4923 VVSGGRGVGSADNFSVVEALADSLGAAVGASRAAVDSGYYPGQFQIGQTG
MAB_3362c|M.abscessus_ATCC_199 VVSGGRGVGSADKFSVVEALADSLGAAVGASRAAVDSGYYPGQFQVGQTG
MAV_3875|M.avium_104 VVSGGRGVGSAENFKVVEELADALGAAVGASRAAVDSGYYPGQFQVGQTG
Mflv_4258|M.gilvum_PYR-GCK VVAGGRGVGSADNFTIVEELADSLGAAVGASRAAVDSGYYPGQFQVGQTG
Mvan_2100|M.vanbaalenii_PYR-1 VVSGGRGVGSAENFGIVEELADSLGGAVGASRAAVDSGYYPGQFQVGQTG
MSMEG_2352|M.smegmatis_MC2_155 VVAGGRGVGSAENFKIVEELADSLGGAVGASRAAVDSGYYPGQFQVGQTG
TH_2195|M.thermoresistible__bu VVAGGRGVGSAENFRVVEELADALGGAVGASRAAVDSGYYPGQFQIGQTG
**:********::* :** ***:**.*******************:****
MMAR_1685|M.marinum_M KTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKDEEAPIFEIADYGVIG
MUL_1923|M.ulcerans_Agy99 KTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKDEEAPIFEIADYGVIG
Mb3054c|M.bovis_AF2122/97 KTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKDEEAPIFEIADYGVVG
Rv3028c|M.tuberculosis_H37Rv KTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKDEEAPIFEIADYGVVG
MLBr_01711|M.leprae_Br4923 KTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKDEEAPIFEIADFGVVG
MAB_3362c|M.abscessus_ATCC_199 KTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKDEEAPIFEIADFGVVG
MAV_3875|M.avium_104 KTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKDEEAPIFEIADYGVVG
Mflv_4258|M.gilvum_PYR-GCK KTVSPQLYIALGISGAIQHRAGMQTSKTIIAVNKDEEAPIFEIADLGIVG
Mvan_2100|M.vanbaalenii_PYR-1 KTVSPQLYIALGISGAIQHRAGMQTSKTIIAVNKDEEAPIFEIADLGIVG
MSMEG_2352|M.smegmatis_MC2_155 KTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKDEEAPIFEIADLGIVG
TH_2195|M.thermoresistible__bu KTVSPQLYIALGISGAIQHRAGMQTSKTIVAVNKDEEAPIFEIADLGIVG
*****************************:*************** *::*
MMAR_1685|M.marinum_M DLFKVAPQLTEGVKARKG
MUL_1923|M.ulcerans_Agy99 DLFKVAPQLTEGVKARKG
Mb3054c|M.bovis_AF2122/97 DLFKVAPQLTEVIKARKG
Rv3028c|M.tuberculosis_H37Rv DLFKVAPQLTEAIKARKG
MLBr_01711|M.leprae_Br4923 DLFKVAPQLTDGIKARKG
MAB_3362c|M.abscessus_ATCC_199 DLFNVAPQLTDAVKARKG
MAV_3875|M.avium_104 DLFKVAPQLTEAIKARKG
Mflv_4258|M.gilvum_PYR-GCK DLFKVTPQLTEAVKARKG
Mvan_2100|M.vanbaalenii_PYR-1 DLFKVTPQLTEAVKARKG
MSMEG_2352|M.smegmatis_MC2_155 DLFKVSPQLTEAVKARKG
TH_2195|M.thermoresistible__bu DLFKVAPQLTEAIKARKG
***:*:****: :*****