For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTFTVDDVAYLRSAAGRQALDEAGKLALSGARLVNDIAALRAAFGDRAAVLAETVQLRRRAAAKFADPSD WLFTDEALQQATAAPVAAHRAARLAGATVHDATCSIGGELVALRDSADHVVGSDLDPVRVAMAAGNVPDV EVCRADALRPITRDAVVILDPARRAGGRRRFDPRAYTPALDAVFDVYRDRDMVVKCAPGIDFAEIERMGF TGEIEVTSVGGGVREACLWSPALAGAGVRRRATVLDRGEQITDTEPDDCAVAPAGRWIVDPDGAVVRAGL VRHYAARHGLWQLDPDIAYLSGDRLPDGVRGFEVLEELGYHERRLRAALAARSAGAVEILVRGVNVDPDV LRTRLRLRGSKQLTVVITRLGSGSASRATAYICRPSR
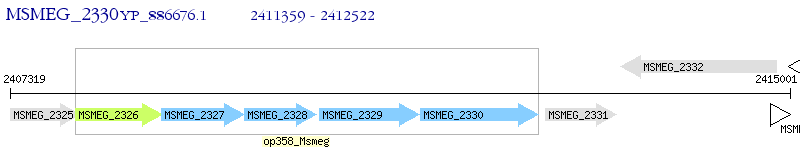
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2330 | - | - | 100% (387) | SAM-dependent methyltransferase |
| M. smegmatis MC2 155 | MSMEG_1050 | - | 0.0 | 100.00% (387) | SAM-dependent methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3063c | - | 1e-136 | 67.88% (358) | hypothetical protein Mb3063c |
| M. gilvum PYR-GCK | Mflv_4268 | - | 1e-152 | 70.43% (372) | hypothetical protein Mflv_4268 |
| M. tuberculosis H37Rv | Rv3037c | - | 1e-137 | 68.16% (358) | hypothetical protein Rv3037c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3380c | - | 1e-134 | 66.06% (386) | hypothetical protein MAB_3380c |
| M. marinum M | MMAR_1664 | - | 1e-132 | 69.86% (345) | hypothetical protein MMAR_1664 |
| M. avium 104 | MAV_3902 | - | 1e-142 | 66.15% (390) | SAM-dependent methyltransferase |
| M. thermoresistible (build 8) | TH_2188 | - | 1e-152 | 74.93% (363) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1901 | - | 1e-132 | 69.57% (345) | hypothetical protein MUL_1901 |
| M. vanbaalenii PYR-1 | Mvan_2091 | - | 1e-160 | 74.80% (381) | hypothetical protein Mvan_2091 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3063c|M.bovis_AF2122/97 ---------------------------------------MRARFGARAPW
Rv3037c|M.tuberculosis_H37Rv ---------------------------------------MRARFGDRAPW
MAV_3902|M.avium_104 -MFSLPDVAYLRSDAGVDALAAVAELELSDATRIADIAAARARFGERTPL
MMAR_1664|M.marinum_M -------------------------------------------------M
MUL_1901|M.ulcerans_Agy99 -------------------------------------------------M
MSMEG_2330|M.smegmatis_MC2_155 MTFTVDDVAYLRSAAGRQALDEAGKLALSGARLVNDIAALRAAFGDRAAV
TH_2188|M.thermoresistible__bu -------------------------MPLTDATRVADVAALRTRFGDRAPV
Mflv_4268|M.gilvum_PYR-GCK -------------------MSEVARYDLTESSLISDIASARKRFGDRTAV
Mvan_2091|M.vanbaalenii_PYR-1 -------MAYLRSDAGRLALAEVSRHRLTDATLVSDIAAARTLFGDRTGV
MAB_3380c|M.abscessus_ATCC_199 -MFDLSDVAYLSSEAGAADLARASTYTFSAGTLVADTAALRNEFGDHAAV
Mb3063c|M.bovis_AF2122/97 LVETTLLRRRAAGKLGELCPNVGVSQWLFTDEALQQATAAPVARHRARRL
Rv3037c|M.tuberculosis_H37Rv LVETTLLRRRAAGKLGELCPNVGVSQWLFTDEALQQATAAPVARHRARRL
MAV_3902|M.avium_104 LVETVLLRRRASEKLGELG----VSDWLFTDEALQQATAAPVAVHRAGRL
MMAR_1664|M.marinum_M LIETTLLRRRAVAKLGGLCPSTPVSDWLFTDTALQQATVAAVALHRARRL
MUL_1901|M.ulcerans_Agy99 LIETTLLRRQAVAKLGGLCPSTPVSDWLFTDTALQQATVAAVALHRARRL
MSMEG_2330|M.smegmatis_MC2_155 LAETVQLRRRAAAKFADPS------DWLFTDEALQQATAAPVAAHRAARL
TH_2188|M.thermoresistible__bu LVETTLLRRRAAAKFDDPS------RWLFTDEALQQATAAPVARHRARRL
Mflv_4268|M.gilvum_PYR-GCK LVETVKLRRKAAVKFVDAG------EWLFTDDALQQATAAPVAAHRARRL
Mvan_2091|M.vanbaalenii_PYR-1 LVETVKLRRKASAKFADAS------GWLFTDDALQQATAAPVAEHRARRL
MAB_3380c|M.abscessus_ATCC_199 LAQTVQLRRKAAAKLGRVD------HWLFTDDALQQATPAAVARHRAARL
* :*. ***:* *: ***** ****** *.** *** **
Mb3063c|M.bovis_AF2122/97 AGR--VVHDATCSIGTELAALRELAVRAVGSDIDPVRLAMARHNLAALGM
Rv3037c|M.tuberculosis_H37Rv AGR--VVHDATCSIGTELAALRELAVRAVGSDIDPVRLAMARHNLAALGM
MAV_3902|M.avium_104 AGPGVVVHDVTCSIGTELAALRARGVTAVGSDIDPVRLAMARHN---IGS
MMAR_1664|M.marinum_M ADL--TVHDATCSIGTELAALCRSAATVIGSDIDPVRLAMARHN---LGG
MUL_1901|M.ulcerans_Agy99 ADL--TVHDATCSIGTELAALCRSAATVIGSDIDPVRLAMARHN---LGG
MSMEG_2330|M.smegmatis_MC2_155 AGA--TVHDATCSIGGELVALRDSADHVVGSDLDPVRVAMAAGNV-P---
TH_2188|M.thermoresistible__bu TGR--KVHDVTCSIGAELAALRLTAAQLVGSDIDPVRLQMARHNLGP---
Mflv_4268|M.gilvum_PYR-GCK AGA--IVHDATCSVGTELAALRNSAALVLGSDIDPVRVAMASHNLGLRGP
Mvan_2091|M.vanbaalenii_PYR-1 AGA--TVHDATCSIGAELAALRNWAGFLLGSDIDPVRLAMARHNLGG---
MAB_3380c|M.abscessus_ATCC_199 AGL--VVHDVTCSVGAELAALDGVAAAVIGSDIDPVRTAMARNN---IGE
:. ***.***:* **.** . :***:**** ** *
Mb3063c|M.bovis_AF2122/97 EADLCRADVLHPVTRDAVVVIDPARRSNGRRRFHLADYQPGLGPLLDRYR
Rv3037c|M.tuberculosis_H37Rv EADLCRADVLHPVTRDAVVVIDPARRSNGRRRFHLADYQPGLGPLLDRYR
MAV_3902|M.avium_104 AAWLCRADALHPVTRDAVLIVDPARRVDGRRRLRIDDYQPGLSPLLDSYR
MMAR_1664|M.marinum_M AVGLCRADALRPVTRDTVVLVDPARRSKGRRRFSPADYQPGLGPVLDVYR
MUL_1901|M.ulcerans_Agy99 AVGLCRADALRPVTRDTVVLVDPARRSKGRRRFSPADYQPGLGPVLDVYR
MSMEG_2330|M.smegmatis_MC2_155 DVEVCRADALRPITRDAVVILDPARRAGGRRRFDPRAYTPALDAVFDVYR
TH_2188|M.thermoresistible__bu GVALCRADALRPITRDTVVVADPARRSGGRRRFDPRAHTPPLDALFDVYR
Mflv_4268|M.gilvum_PYR-GCK GVALCRADALRPVSEGAVVLVDPARRSGSRRRFDPRAYLPPLDTLLDVLR
Mvan_2091|M.vanbaalenii_PYR-1 SVPLCRADALRPITVDAVVVVDPARRTGSRRRFDPRSYLPPLDDLLEVLR
MAB_3380c|M.abscessus_ATCC_199 HVLVCRADALAPSSRAQVVIADPGRRAGGRRRFDPSAYSPPLDAVLRSYA
. :****.* * : *:: **.** .***: : * *. ::
Mb3063c|M.bovis_AF2122/97 GRDVVVKCAPGIDFEEV-GRLGFEGEIEVISYRGGVREACLWSAGLAGSG
Rv3037c|M.tuberculosis_H37Rv GRDVVVKCAPGIDFEEV-GRLGFEGEIEVISYRGGVREACLWSAGLAGSG
MAV_3902|M.avium_104 GRDFVVKCAPGIDFEQV-RRLGFAGEIEVTSYRGSVREACLWSAGLTQSG
MMAR_1664|M.marinum_M DRPLVVKCAPGIDFDQV-SRLGFDGEIEVTSYQGSVREACLWSRGLAAAG
MUL_1901|M.ulcerans_Agy99 DRPLVVKCAPGIDFDQV-SRLGFDGEIEVTSYQGSVREACLWSRGLAAAG
MSMEG_2330|M.smegmatis_MC2_155 DRDMVVKCAPGIDFAEI-ERMGFTGEIEVTSVGGGVREACLWSPALAGAG
TH_2188|M.thermoresistible__bu GRDSVVKCAPGIEFGAL-GAVGFDGEVEVVSLAGSVREACLWSGSLTEPG
Mflv_4268|M.gilvum_PYR-GCK ERDYVVKCAPGIDFAVV-GELGFRGEIELVSLGGSVREACLWSSGFAADG
Mvan_2091|M.vanbaalenii_PYR-1 DRHHVVKCAPGIDFAVL-EQLGFDGEIEVTSLSGSVREACLWSRGLSDGG
MAB_3380c|M.abscessus_ATCC_199 DRDLVVKCAPGLDFADLREQTGFDGEIEVVSLDGGVREACLWSAGLA--G
* *******::* : ** **:*: * *.******** .:: *
Mb3063c|M.bovis_AF2122/97 IRRRASILD---SGEQIGDDEPDDCGVRPAGKWIVDPDGAVVRAGLVRNY
Rv3037c|M.tuberculosis_H37Rv IRRRASILD---SGEQIGDDEPDDCGVRPAGKWIVDPDGAVVRAGLVRNY
MAV_3902|M.avium_104 VRRRATVLD---RGEQITDTDPDDCPVRPVGRWIVDPDGAVVRAGLVRHY
MMAR_1664|M.marinum_M VRRRASILD---SGEQITDSEPDDCDVRPAGRWIIDPDGAVVRAGLVRHY
MUL_1901|M.ulcerans_Agy99 VRRRASILD---SGEQITDSEPDDCDVRPAGRWIIDPDGAVVRAGLVRHY
MSMEG_2330|M.smegmatis_MC2_155 VRRRATVLD---RGEQITDTEPDDCAVAPAGRWIVDPDGAVVRAGLVRHY
TH_2188|M.thermoresistible__bu VRRRATILD---RSEVITDAEPGECPVAAAGRWIIDPDGAVVRAGLVRHY
Mflv_4268|M.gilvum_PYR-GCK VRRRATMLD---TGEQVTDTDPGQCPVAPAGRWIVDPDGAVVRAGLVRHY
Mvan_2091|M.vanbaalenii_PYR-1 VRRRATLLD---TGEQLTDTEADGCAVAPAGRWIIDPDGAVVRSGLVRHY
MAB_3380c|M.abscessus_ATCC_199 ARRRATVLATIGTGYQLTDADPDDCAVAGAGEWIIDPDGAVVRAGLVRQY
****::* . : * :.. * * .*.**:********:****:*
Mb3063c|M.bovis_AF2122/97 GARHGLWQLDPQIAYLSGDRLPPALRGFEVLEQLAFDERRLRQVLSALDC
Rv3037c|M.tuberculosis_H37Rv GARHGLWQLDPQIAYLSGDRLPPALRGFEVLEQLAFDERRLRQVLSALDC
MAV_3902|M.avium_104 GARHELWQLDPDIAYLSGDRLPATVRGLRVLEQLAFDERRLRQALTASDC
MMAR_1664|M.marinum_M GARHRLWQLDPNIAYLSGDTLPEGIRGFEVLEALPFDERRLRQALNALDC
MUL_1901|M.ulcerans_Agy99 GARHRLWQLDPNIAYLSGDTLPEGIRGFEVLEALPFDERRLRQALNALDC
MSMEG_2330|M.smegmatis_MC2_155 AARHGLWQLDPDIAYLSGDRLPDGVRGFEVLEELGYHERRLRAALAARSA
TH_2188|M.thermoresistible__bu AARHGLWQLDPDIAYLSGDRLPAGVRGFEVLEELPYRERRLRQALAAHDA
Mflv_4268|M.gilvum_PYR-GCK AARHGLWQLDPDIAYLSGDRLPDGVRGFEVLDQVAFSERHLRQALSALDA
Mvan_2091|M.vanbaalenii_PYR-1 AARHGLWQLDPDIAYLSGDRLPPGVRGFEVMEQLAFGERQLRQALSARDA
MAB_3380c|M.abscessus_ATCC_199 AARHGLWQLDARIAYLSGDCVPAGVQGYRVLDSLPYSEKRLRQALAARDC
.*** *****. ******* :* ::* .*:: : : *::** .* * ..
Mb3063c|M.bovis_AF2122/97 GAAEILVRGVAIDPDALRRRLRLRGSRPLAVVITRIGAGS-----LSHVT
Rv3037c|M.tuberculosis_H37Rv GAAEILVRGVAIDPDALRRRLRLRGSRPLAVVITRIGAGS-----LSHVT
MAV_3902|M.avium_104 GALEVLVRGVRVDPDALRKRMRLRGSRPLSVVITRIGART-----SGQVT
MMAR_1664|M.marinum_M GALEILVRGVQVDPDALRRRLRPRGGRALSVVITRIGTGS-----ASHVT
MUL_1901|M.ulcerans_Agy99 GALEILVRGVQVDPDALRRRLRPRGGRALSVVITRIGTGS-----ASHVT
MSMEG_2330|M.smegmatis_MC2_155 GAVEILVRGVNVDPDVLRTRLRLRGSKQLTVVITRLGSGS-----ASRAT
TH_2188|M.thermoresistible__bu GAVEILVRGVDVDPDRLRTRLRLRGRHPLAVVITRIGSKH-----ASRAT
Mflv_4268|M.gilvum_PYR-GCK GALEILVRGLDVDPDALRARLRPRGAAQISVVVTRIGSGA-----ASRAT
Mvan_2091|M.vanbaalenii_PYR-1 GAVEILVRGVDVDPDVLRSRLRLRGSAQVTVVIARLGSGV-----ASRAT
MAB_3380c|M.abscessus_ATCC_199 GAVEITVRGVDLDPALLRTRLKLRGSVPLSVVITRIGSGAKAFVCAARSP
** *: ***: :** ** *:: ** ::**::*:*: .: .
Mb3063c|M.bovis_AF2122/97 AYVCRPSR---
Rv3037c|M.tuberculosis_H37Rv AYVCRPSR---
MAV_3902|M.avium_104 AFVCQPSR---
MMAR_1664|M.marinum_M AYVCCPSR---
MUL_1901|M.ulcerans_Agy99 AYVCCPSR---
MSMEG_2330|M.smegmatis_MC2_155 AYICRPSR---
TH_2188|M.thermoresistible__bu AFICRPSR---
Mflv_4268|M.gilvum_PYR-GCK AFLCRPSR---
Mvan_2091|M.vanbaalenii_PYR-1 AFICRPSR---
MAB_3380c|M.abscessus_ATCC_199 GGTRTPDPRSA
. *.